I also searched without success for multivariate KDE conditional sampling functionality, so I wrote a subclass of scipy.stats.gaussian_kde() with a conditional_resample() method: https://pypi.org/project/kdetools/
You can conditionally sample from any subset of the variables, do vectorised sampling for multiple conditioning vectors, as well as doing bandwidth selection using k-fold CV using different kinds of bandwidth matrices. The code is still somewhat a work in progress, e.g. need to add some tests, and I can't quite replicate statsmodels/R multivariate bandwidth selection results. Happy for people to stress-test it and give feedback.
As a toy example, I created a 3-component Gaussian mixture, sampled from it and fit KDEs using a few different bandwidth approaches:
import numpy as np
import matplotlib.pyplot as plt
import kdetools as kt
# Generate toy 2D mixture of 3 Gaussians, all centred on 0 in the x-direction
np.random.seed(0)
N = 200
X = np.vstack([np.random.multivariate_normal([0,0],[[1.5,-0.5],[-0.5,1]], size=N),
np.random.multivariate_normal([0,3],[[1,0.5],[0.5,1.5]], size=N),
np.random.multivariate_normal([0,6],[[2,1],[1,1.5]], size=N)])
# Define the mixture pdf
def zz_pdf_func(X):
zz_pdf1 = st.multivariate_normal([0,0],[[1.5,-0.5],[-0.5,1]])
zz_pdf2 = st.multivariate_normal([0,3],[[1,0.5],[0.5,1.5]])
zz_pdf3 = st.multivariate_normal([0,6],[[2,1],[1,1.5]])
return (zz_pdf1.pdf(X)+zz_pdf2.pdf(X)+zz_pdf3.pdf(X))/3
# Fit KDEs using different bandwidth types
kde1 = kt.gaussian_kde(X.T)
kde2 = kt.gaussian_kde(X.T); kde2.set_bandwidth('cv', bw_type='equal')
kde3 = kt.gaussian_kde(X.T); kde3.set_bandwidth('cv', bw_type='diagonal')
kde4 = kt.gaussian_kde(X.T); kde4.set_bandwidth('cv', bw_type='covariance')
# Plot 2D KDEs and overlay contours of true distribution in red:
xx, yy = np.meshgrid(np.linspace(-5, 5, 1000), np.linspace(-3, 10, 1000))
zz_pdf = zz_pdf_func(np.stack([xx.ravel(), yy.ravel()]).T).reshape(xx.shape)
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2, figsize=(10,10))
zz_kde1 = kde1.pdf(np.stack([xx.ravel(), yy.ravel()])).reshape(xx.shape)
zz_kde2 = kde2.pdf(np.stack([xx.ravel(), yy.ravel()])).reshape(xx.shape)
zz_kde3 = kde3.pdf(np.stack([xx.ravel(), yy.ravel()])).reshape(xx.shape)
zz_kde4 = kde4.pdf(np.stack([xx.ravel(), yy.ravel()])).reshape(xx.shape)
ax1.contourf(xx, yy, zz_kde1, alpha=0.95)
ax1.contour(xx, yy, zz_pdf, colors='r', linewidths=0.25)
ax1.scatter(X[:,0], X[:,1], s=1)
ax1.set_title('Standard - scaled full covariance')
ax2.contourf(xx, yy, zz_kde2, alpha=0.95)
ax2.contour(xx, yy, zz_pdf, colors='r', linewidths=0.25)
ax2.scatter(X[:,0], X[:,1], s=1)
ax2.set_title('CV - spherical covariance matrix')
ax3.contourf(xx, yy, zz_kde3, alpha=0.95)
ax3.contour(xx, yy, zz_pdf, colors='r', linewidths=0.25)
ax3.scatter(X[:,0], X[:,1], s=1)
ax3.set_title('CV - diagonal covariance matrix')
ax4.contourf(xx, yy, zz_kde4, alpha=0.95)
ax4.contour(xx, yy, zz_pdf, colors='r', linewidths=0.25)
ax4.scatter(X[:,0], X[:,1], s=1)
ax4.set_title('CV - scaled full covariance matrix');
![KDEs generated from Gaussian mixture]() Having generated the KDE, I took conditional samples of
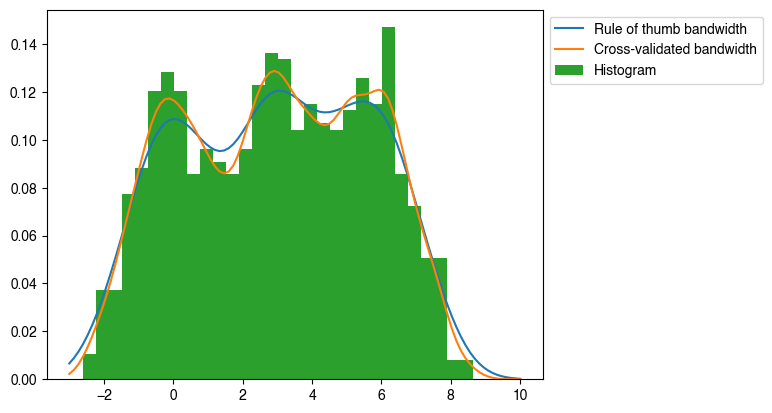
Having generated the KDE, I took conditional samples of y|x=0 and generated another histogram/KDE:
np.random.seed(42)
y = kde3.conditional_resample(1000, x_cond=np.array([0]), dims_cond=[0]).ravel()
kde1d = kt.gaussian_kde(y)
xs = np.linspace(-3, 10, 100)
plt.plot(xs, kde1d.pdf(xs), label='Rule of thumb bandwidth')
# Update the bandwidth
kde1d.set_bandwidth(bw_method='cv', bw_type='equal')
xs = np.linspace(-3, 10, 100)
plt.plot(xs, kde1d.pdf(xs), label='Cross-validated bandwidth')
plt.hist(y, bins=30, density=True, label='Histogram')
plt.legend(bbox_to_anchor=(1,1));
![Conditional samples from KDE]()