I took the matlab code from this tutorial Texture Segmentation Using Gabor Filters.
To test clustering algorithms on the resulting multi-dimensional texture responses to gabor filters, I applied Gaussian Mixture and Fuzzy C-means instead of the K-means to compare their results (number of clusters = 2 in all of the cases):
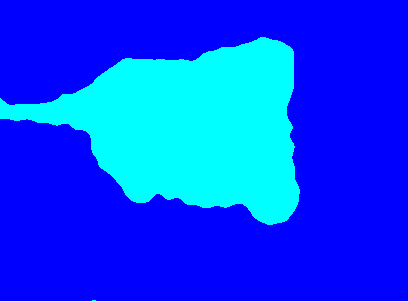
Original image:
K-means clusters:
L = kmeans(X, 2, 'Replicates', 5);
GMM clusters:
options = statset('MaxIter',1000);
gmm = fitgmdist(X, 2, 'Options', options);
L = cluster(gmm, X);
Fuzzy C-means:
[centers, U] = fcm(X, 2);
[values indexes] = max(U);
What I've found weird in this case is that K-means clusters are more accurate than those extracted using GMM and Fuzzy C-means.
Can anyone explain to me if the high-dimensionality (L x W x 26: 26 is the number of gabor filters used) of the data given as input to the GMM and the Fuzzy C-means classifiers is what's causing the clustering to be less accurate?
In other words is the GMM and the Fuzzy C-means clustering more sensitive to the dimensionality of the data, than K-means is?




'Replicates'and higher quality initialization (k-means++). k-means isGMMunder a spherical-covariance assumption, so in theory it shouldn't do much better. I think most of the discrepancy comes down to initialization. You should be able to test this by using the k-means result as initial conditions forGMM. – Konya