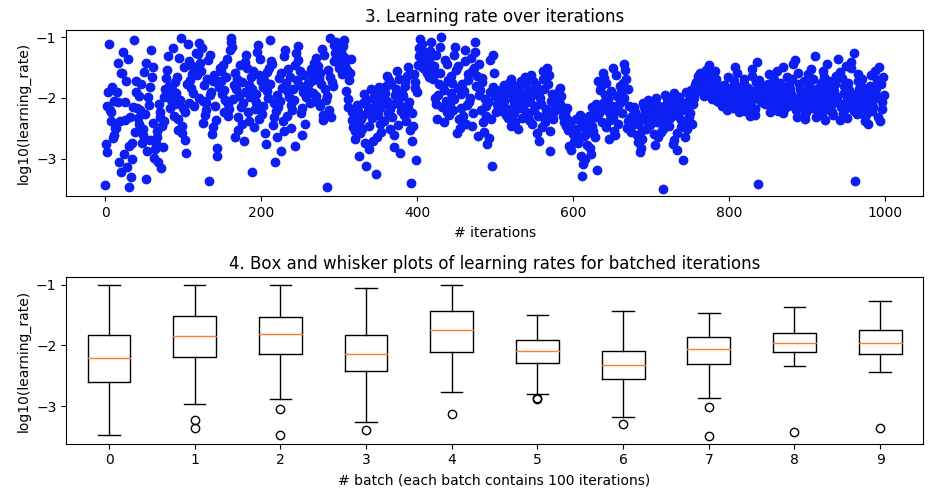
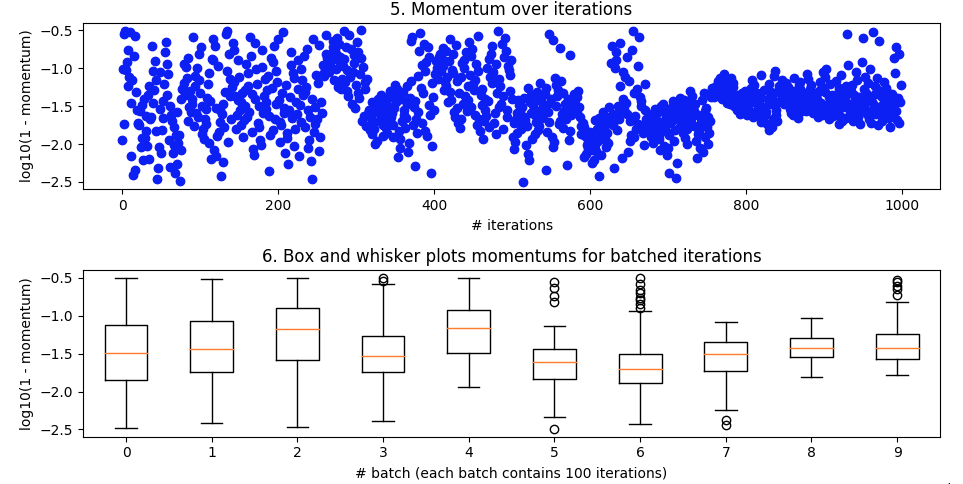
I tried to apply Bayesian Optimisation to a simple CNN for the MNIST handwritten digits dataset and I'm getting little indication that it works. I've tried doing k-fold validation to smooth out noise but still doesn't seem like the optimisation is making any headway in converging towards optimal parameters. In general, what are some of the main reasons Bayesian Optimization might fail? And in my particular case?
The rest of this is just context and code snippets.
Model definition:
def define_model(learning_rate, momentum):
model = Sequential()
model.add(Conv2D(32, (3,3), activation = 'relu', kernel_initializer = 'he_uniform', input_shape=(28,28,1)))
model.add(MaxPooling2D((2,2)))
model.add(Flatten())
model.add(Dense(100, activation='relu', kernel_initializer='he_uniform'))
model.add(Dense(10, activation='softmax'))
opt = SGD(lr=learning_rate, momentum=momentum)
model.compile(optimizer=opt, loss='categorical_crossentropy', metrics=['accuracy'])
return model
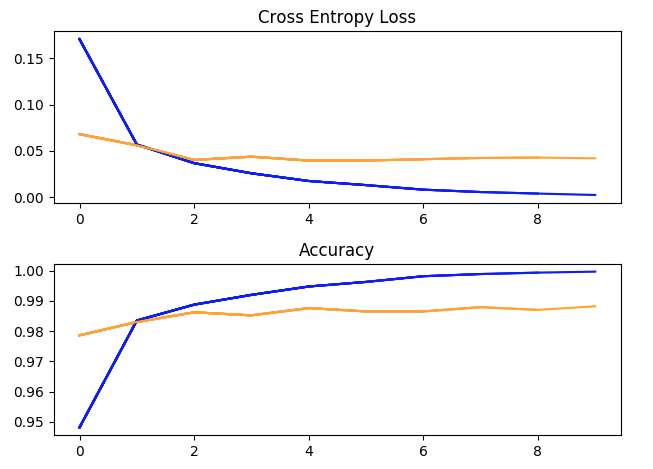
One training run with hyperparameters: batch_size = 32, learning rate = 1e-2, momentum = 0.9, 10 epochs. (blue = training, yellow = validation).
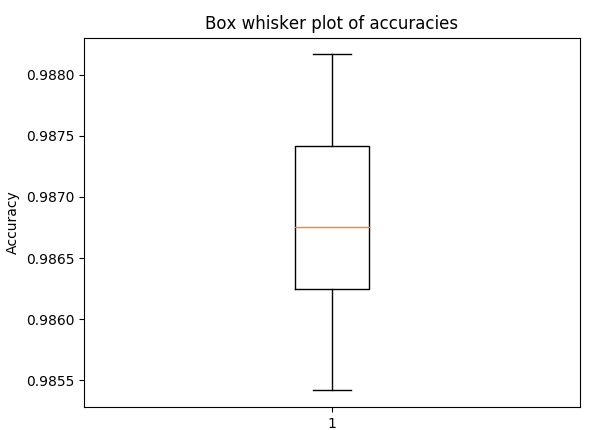
Box and whisker plot for accuracy in 5-fold cross validation, with the same hyperparameters as above (to get a sense of the spread)
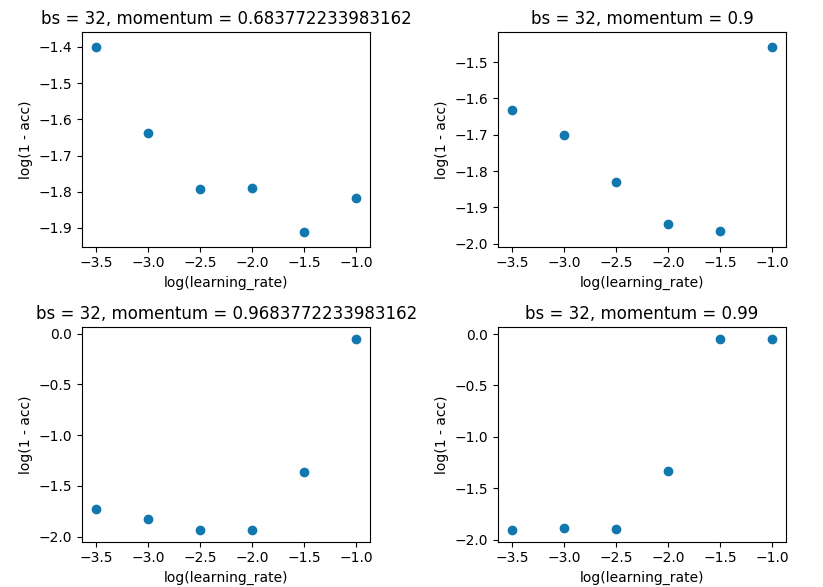
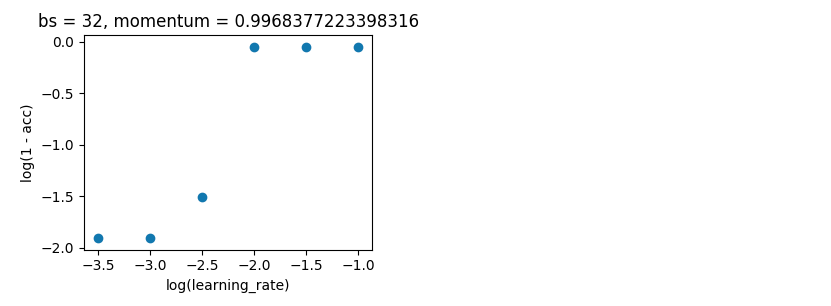
Grid search holding batch_size at 32, and keeping with 10 epoch. I did this on single evaluations rather than 5-fold as the spread wasn't large enough to spoil the results.
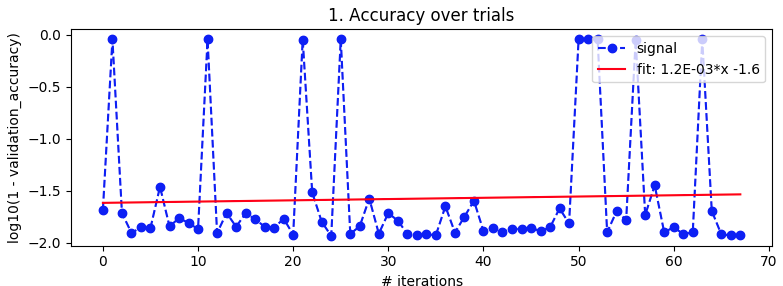
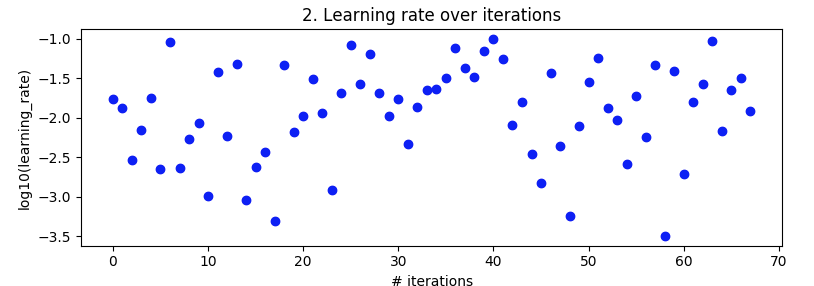
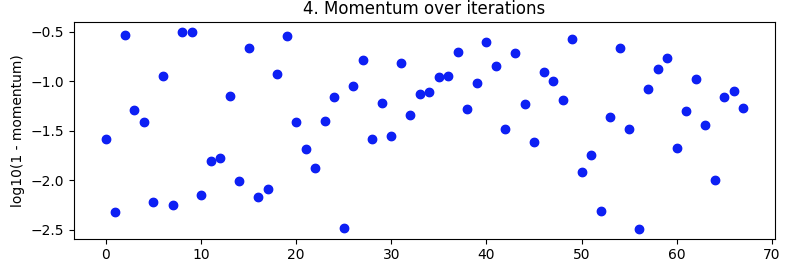
Bayesian optimization. As above, batch_size=32 and 10 epoch. Searching over the same ranges. But this time with 5-fold cross-validation to smooth out noise. It's supposed to go out to 100 iterations but that's still another 20 hours away.
space = {'lr': hp.loguniform('lr', np.log(np.sqrt(10)*1e-4), np.log(1e-1)), 'momentum': 1 - hp.loguniform('momentum', np.log(np.sqrt(10)*1e-3), np.log(np.sqrt(10)*1e-1))}
tpe_best = fmin(fn=objective, space=space, algo=tpe.suggest, trials=Trials(), max_evals=100)
It looked nice from about iteration 27 to 49 but then it lost its mind again.
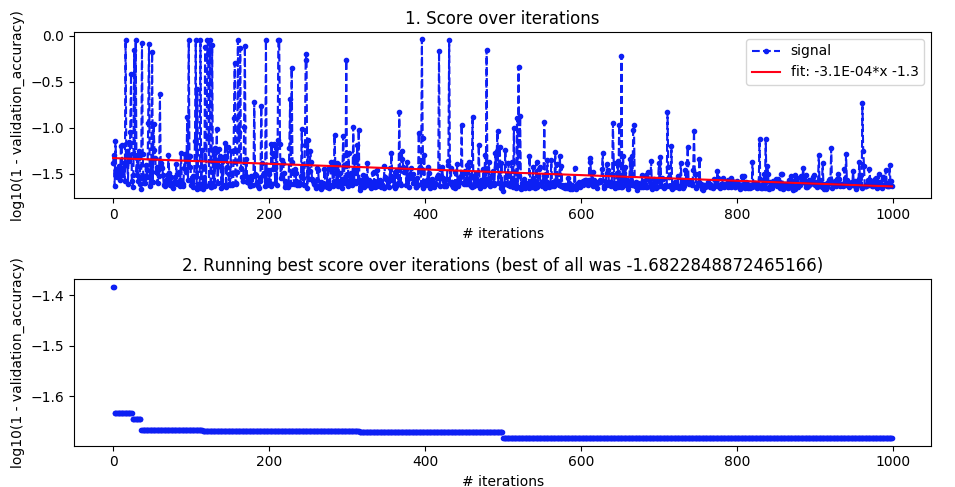
EDIT
More detail for those who asked.
Imports
# basic utility libraries
import numpy as np
import pandas as pd
import time
import datetime
import pickle
from matplotlib import pyplot as plt
%matplotlib notebook
# keras
from keras.datasets import mnist
from keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, Input, BatchNormalization
from keras.optimizers import SGD
from keras.callbacks import Callback
from keras.models import load_model
# learning and optimisation helper libraries
from sklearn.model_selection import KFold
from hyperopt import fmin, tpe, Trials, hp, rand
from hyperopt.pyll.stochastic import sample
Single evaluation
def evaluate_model(trainX, trainY, testX, testY, max_epochs, learning_rate, momentum, batch_size, model=None, callbacks=[]):
if model == None:
model = define_model(learning_rate, momentum)
history = model.fit(trainX, trainY, epochs=max_epochs, batch_size=batch_size, validation_data=(testX, testY), verbose=0, callbacks = callbacks)
return model, history
Cross validation
def evaluate_model_cross_validation(trainX, trainY, max_epochs, learning_rate, momentum, batch_size, n_folds=5):
scores, histories = list(), list()
# prepare cross validation
kfold = KFold(n_folds, shuffle=True, random_state=1)
# enumerate splits
for trainFold_ix, testFold_ix in kfold.split(trainX):
# select rows for train and test
trainFoldsX, trainFoldsY, testFoldX, testFoldY = trainX[trainFold_ix], trainY[trainFold_ix], trainX[testFold_ix], trainY[testFold_ix]
# fit model
model = define_model(learning_rate, momentum)
history = model.fit(trainFoldsX, trainFoldsY, epochs=max_epochs, batch_size=batch_size, validation_data=(testFoldX, testFoldY), verbose=0)
# evaluate model
_, acc = model.evaluate(testFoldX, testFoldY, verbose=0)
# stores scores
scores.append(acc)
histories.append(history)
return scores, histories
How I set things up for Bayesian Optimisation (or random search)
def selective_search(kind, space, max_evals, batch_size=32):
trainX, trainY, testX, testY = prep_data()
histories = list()
hyperparameter_sets = list()
scores = list()
def objective(params):
lr, momentum = params['lr'], params['momentum']
accuracies, _ = evaluate_model_cross_validation(trainX, trainY, max_epochs=10, learning_rate=lr, momentum=momentum, batch_size=batch_size, n_folds=5)
score = np.log10(1 - np.mean(accuracies))
scores.append(score)
with open('{}_scores.pickle'.format(kind), 'wb') as file:
pickle.dump(scores, file)
hyperparameter_sets.append({'learning_rate': lr, 'momentum': momentum, 'batch_size': batch_size})
with open('{}_hpsets.pickle'.format(kind), 'wb') as file:
pickle.dump(hyperparameter_sets, file)
return score
if kind == 'bayesian':
tpe_best = fmin(fn=objective, space=space, algo=tpe.suggest, trials=Trials(), max_evals=max_evals)
elif kind == 'random':
tpe_best = fmin(fn=objective, space=space, algo=rand.suggest, trials=Trials(), max_evals=max_evals)
else:
raise BaseError('First parameter "kind" must be either "bayesian" or "random"')
return histories, hyperparameter_sets, scores
Then how I actually run the Bayesian optimization.
space = {'lr': hp.loguniform('lr', np.log(np.sqrt(10)*1e-4), np.log(1e-1)), 'momentum': 1 - hp.loguniform('momentum', np.log(np.sqrt(10)*1e-3), np.log(np.sqrt(10)*1e-1))}
histories, hyperparameter_sets, scores = selective_search(kind='bayesian', space=space, max_evals=100, batch_size=32)