I am trying to identify the indices of the masked pixels when using maskoceans so I can then call only the land pixels in a code I have that is currently going through the whole globe, even though I do not care about the ocean pixels. I was trying different methods to do so, and noticed that my plots were looking really weird. Eventually, I realized that something was getting mixed up in my lat/lon indices, even though I am not actually touching them! Here is the code:
import numpy as np
import netCDF4
from datetime import datetime, timedelta
import matplotlib
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
import matplotlib.dates as mpldates
import heat_transfer_coeffs
from dew_interface import get_dew
from matplotlib.dates import date2num, num2date
import numpy as np
import netCDF4
import heat_transfer_coeffs as htc
from jug.task import TaskGenerator
import matplotlib.cm as cm
import mpl_toolkits
from mpl_toolkits import basemap
from mpl_toolkits.basemap import Basemap, maskoceans
np.seterr(all='raise')
# set global vars
ifile = netCDF4.Dataset('/Users/myfile.nc', 'r')
times = ifile.variables['time'][:].astype(np.float64) # hours since beginning of dataset
lats_1d = ifile.variables['latitude'][:] # 90..-90
lons_1d = ifile.variables['longitude'][:] # 0..360
lons_1d[lons_1d>180]-=360 #putting longitude into -180..180
lons, lats = np.meshgrid(lons_1d, lats_1d)
ntimes, nlats, nlons = ifile.variables['tm'].shape
ifile.close()
map1 = basemap.Basemap(resolution='c', projection='mill',llcrnrlat=-36 , urcrnrlat=10, llcrnrlon=5 , urcrnrlon=52)
#Mask the oceans
new_lon = maskoceans(lons,lats,lons,resolution='c', grid = 10)
new_lat = maskoceans(lons,lats,lats,resolution='c', grid = 10)
fig = plt.figure
pc = map1.pcolormesh(lons, lats, new_lat, vmin=0, vmax=34, cmap=cm.RdYlBu, latlon=True)
plt.show()
for iii in range(new_lon.shape[1]):
index = np.where(new_lon.mask[:,iii] == False)
index2 = np.where(new_lon.mask[:,iii] == True)
new_lon[index[0],iii] = 34
new_lon[index2[0],iii] = 0
fig = plt.figure
pc = map1.pcolormesh(lons, lats, new_lat, vmin=0, vmax=34, cmap=cm.RdYlBu, latlon=True)
plt.show()
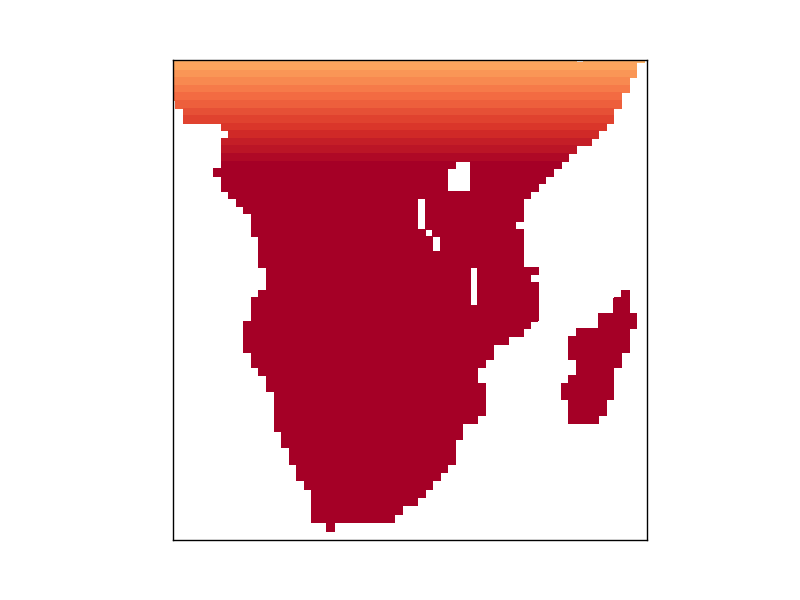
The first figure I get shows the expected map of Africa with oceans masked and the land values corresponding to the latitude (until saturation of the colorbar at 34, but that value was just taken as an example)
However, the second figure, which should plot the exact same thing as the first one, comes out all messed up, even though the loop in between the first and second figure doesn't touch any of the parameters involved in plotting it:
If I comment out the loop in between figure 1 and 2, figure 2 looks just like figure 1. Any idea about what is going on here?

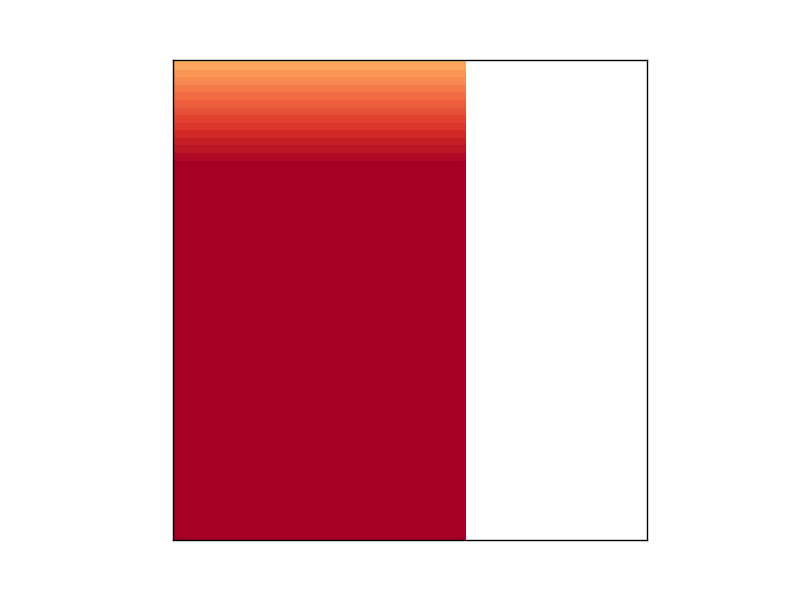
maskoceans...! – Bacchus