The documentation for numpy.linalg.det states that
The determinant is computed via LU factorization using the LAPACK routine z/dgetrf.
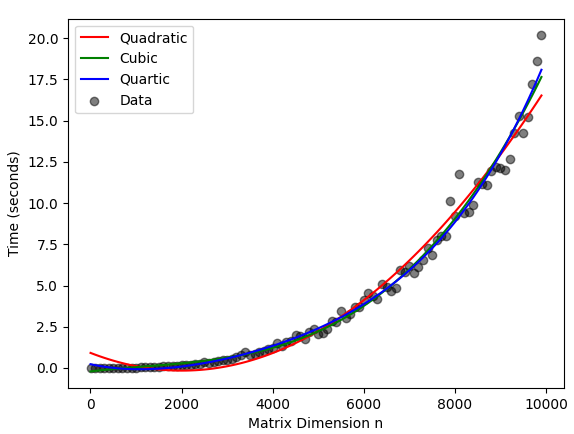
I ran the following run time tests and fit polynomials of degrees 2, 3, and 4 because that covers the least worst options in this table. That table also mentions that an LU decomposition approach takes $O(n^3)$ time, but then the theoretical complexity of LU decomposition given here is $O(n^{2.376})$. Naturally the choice of algorithm matters, but I am not sure what available time complexities I should expect from numpy.linalg.det.
from timeit import timeit
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import PolynomialFeatures
sizes = np.arange(1,10001, 100)
times = []
for size in sizes:
A = np.ones((size, size))
time = timeit('np.linalg.det(A)', globals={'np':np, 'A':A}, number=1)
times.append(time)
print(size, time)
sizes = sizes.reshape(-1,1)
times = np.array(times).reshape(-1,1)
quad_sizes = PolynomialFeatures(degree=2).fit_transform(sizes)
quad_times = LinearRegression().fit(quad_sizes, times).predict(quad_sizes)
cubic_sizes = PolynomialFeatures(degree=3).fit_transform(sizes)
cubic_times = LinearRegression().fit(cubic_sizes, times).predict(cubic_sizes)
quartic_sizes = PolynomialFeatures(degree=4).fit_transform(sizes)
quartic_times = LinearRegression().fit(quartic_sizes, times).predict(quartic_sizes)
plt.scatter(sizes, times, label='Data', color='k', alpha=0.5)
plt.plot(sizes, quad_times, label='Quadratic', color='r')
plt.plot(sizes, cubic_times, label='Cubic', color='g')
plt.plot(sizes, quartic_times, label='Quartic', color='b')
plt.xlabel('Matrix Dimension n')
plt.ylabel('Time (seconds)')
plt.legend()
plt.show()
The output of the above is given as the following plot.
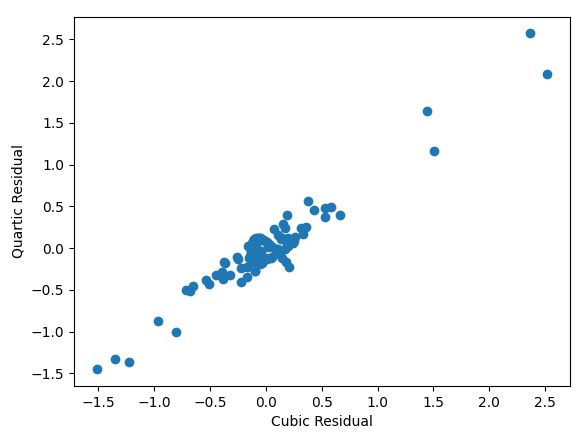
Since none of the available complexities get down to quadratic time, I am unsurprising that visually the quadratic model had the worst fit. Both the cubic and quartic models had excellent visual fit, and unsurprisingly their residuals are closely correlated.
Some related questions exist, but they do not have an answer for this specific implementation.
- Space complexity of matrix inversion, determinant and adjoint
- Time and space complexity of determinant of a matrix
- Experimentally determining computing complexity of matrix determinant
Since this implementation is used by a lot of Python programmers world-wide, it may benefit the understanding of a lot of people if an answer was tracked down.

det()in Numpy is close to O(n^3). Great experiment! – Barrick