I am trying to create a map containing polygons that represent 25km buffer into the ocean from every country's coastline, so that I can calculate the area within this buffer for each country. I am having trouble doing this both with an uploaded coastline file, and with naturalearth data. Here's what I've tried so far:
library(rgdal)
library(sf)
library(rnaturalearthdata)
library(rnaturalearth)
library(rgeos)
library(maps)
library(countrycode)
# World polygons from the maps package
world_shp <- sf::st_as_sf(maps::map("world", plot = T, fill = TRUE))
world_shp_df <- world_shp %>%
mutate(Alpha.3.code = countrycode(ID, "country.name", "iso3c", warn = FALSE))
# Get coastlines using rnaturalearth
coastline <- ne_coastline(scale = "medium", returnclass = "sf")
# Buffer the coastlines by 25 km
buffer_distance <- 25000 # 25 km in meters
coastline_buffers <- st_buffer(coastline, dist = buffer_distance) %>%
st_make_valid()
ggplot() +
geom_sf(data = coastline_buffers , fill = "lightblue")
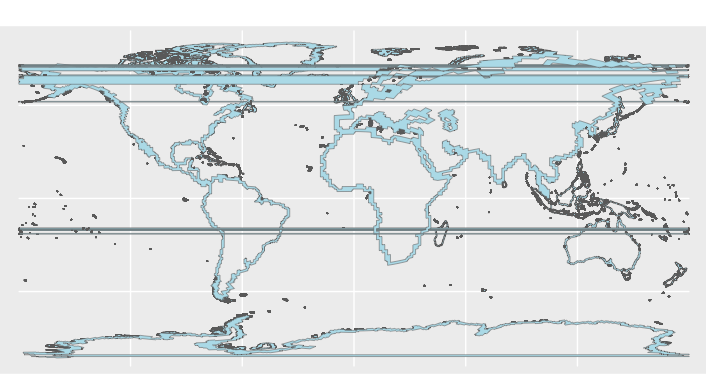
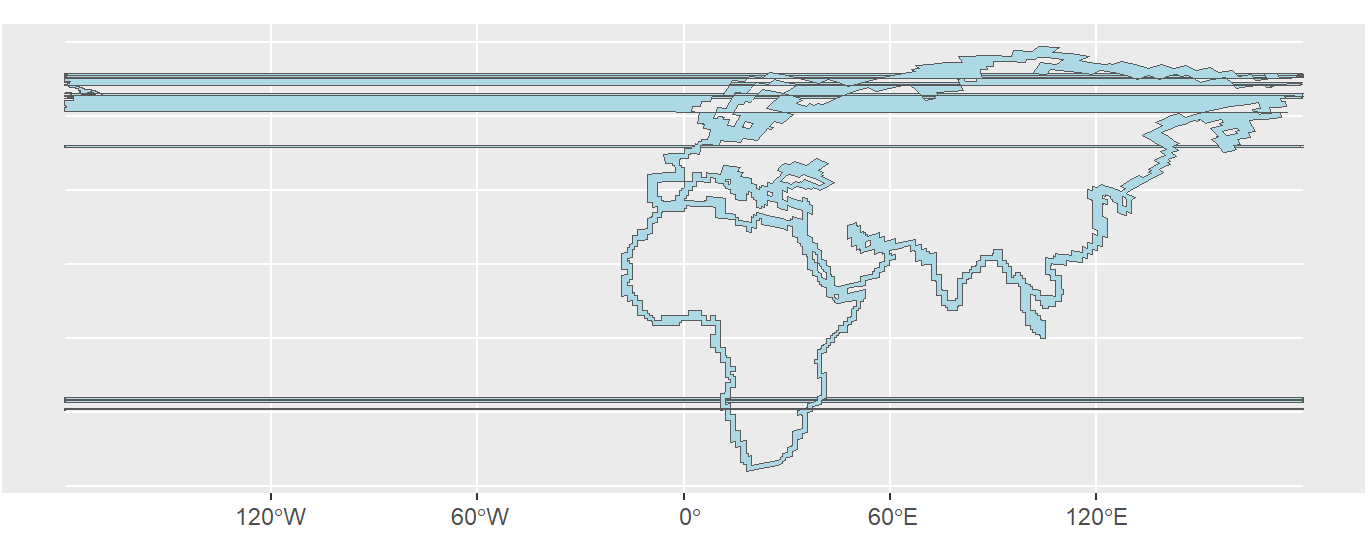
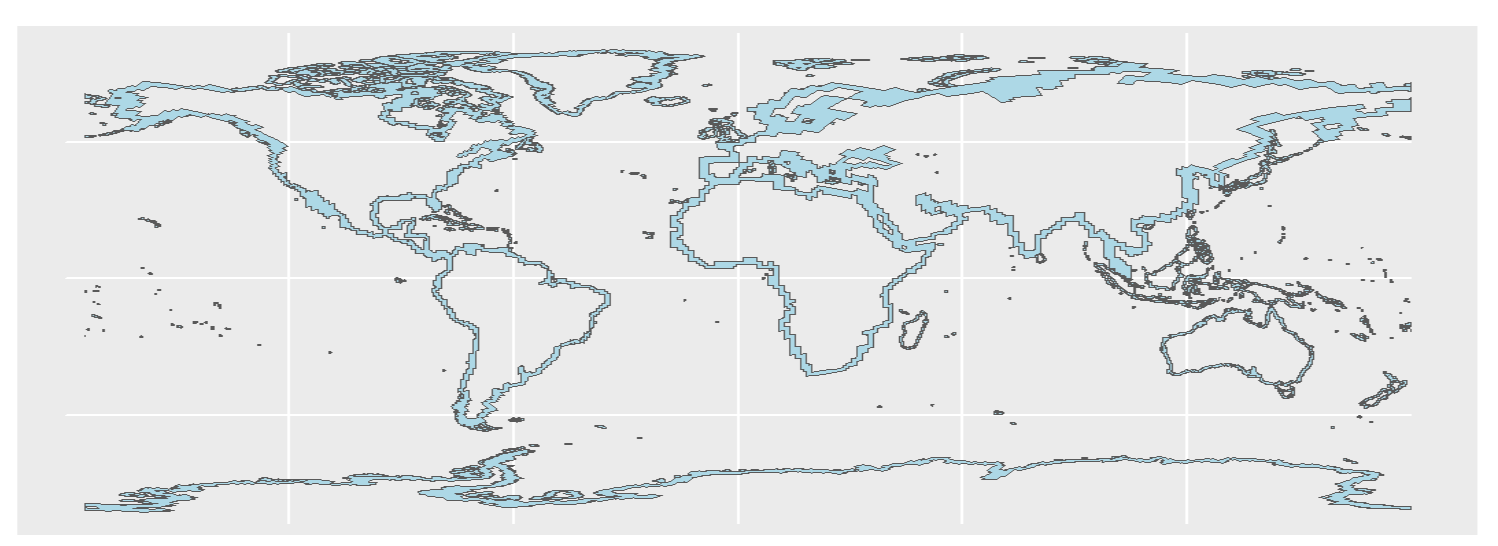
This results in a map with horizontal lines running across it, see the image:
I have tried simplifying the geometry, using a different crs, and I just can't seem to figure it out. Any help would be really appreciated!

rgeosandspatialEcofunctions. – Godden