I am trying to model a three-level nested linear mixed effects model in rjags
(by three-level: multiple observations for multiple individuals within multiple groups).
There are unique sets of individuals in the groups.
The equivalent model in lme4 would be
lmer(yN ~ x + (1 |group/indiv), data=qq)
or
lmer(yN ~ x + (1 |group) + (1|indiv), data=qq)
My question is: How do I program this model in rjags please.
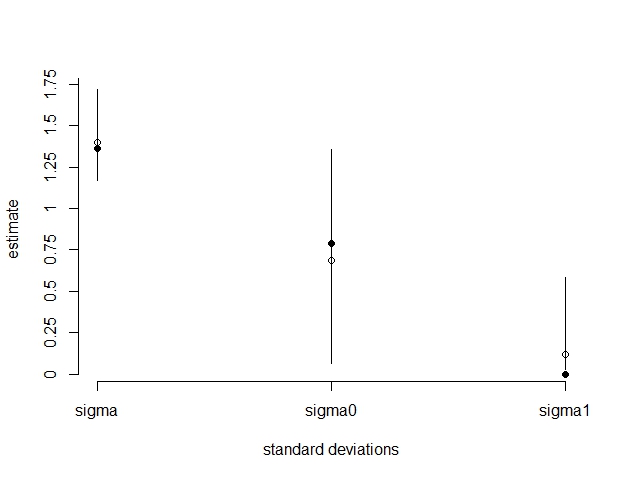
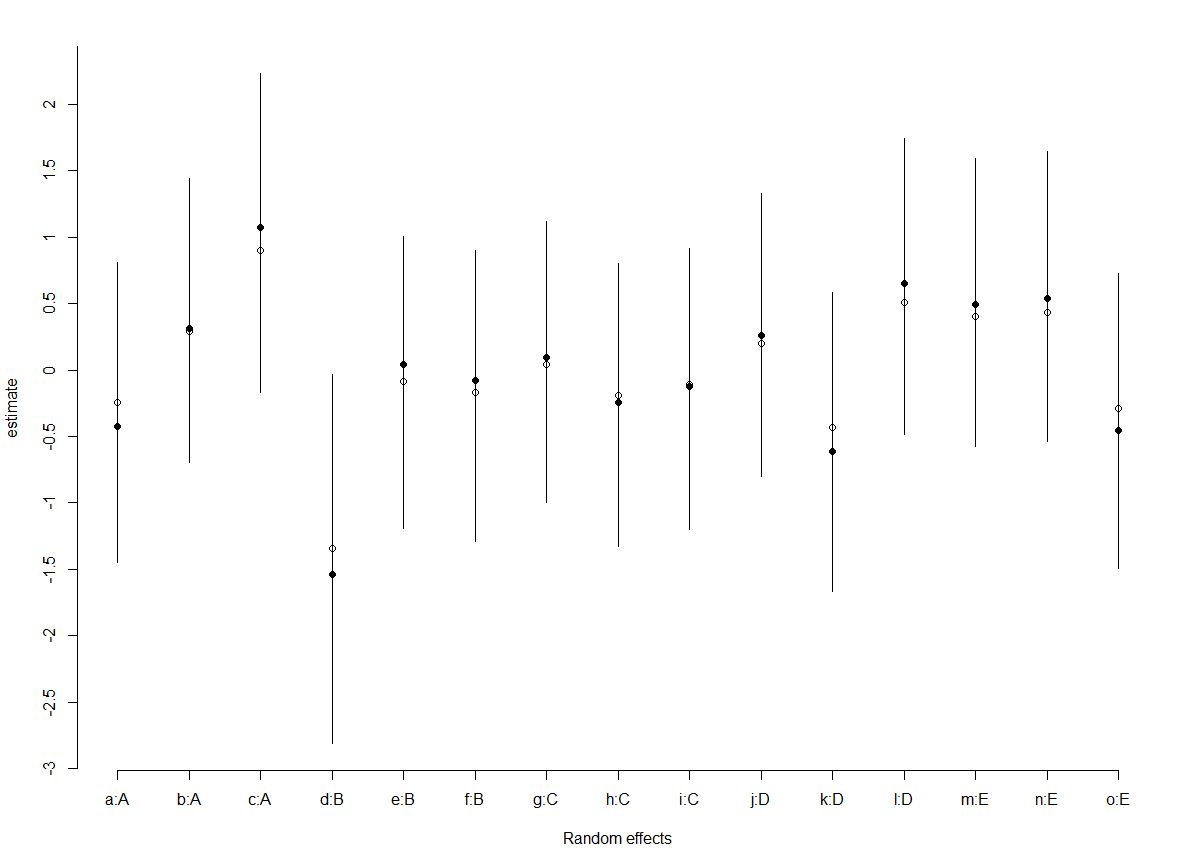
This is my attempt at the rjags code, which compiles, and executes but the individual level random effects seem to be penalised too much - enough to suggest that it is coded incorrectly.
st <- "
model {
for(i in 1:n){
mu[i] <- beta[1] + b1[ind[i]] + b2[group[i]] + beta[2]* x[i]
y[i] ~ dnorm(mu[i], tau)
}
for(i in 1:2){ beta[i] ~ dnorm(0, 0.0001) }
tau ~ dgamma(0.01, 0.01)
sigma <- sqrt(1/tau)
# hierarchical model
for (i in 1:nInd) { b1[i] ~ dnorm(0, tau0) }
for (i in 1:nGrp) { b2[i] ~ dnorm(0, tau1) }
tau0 ~ dgamma(0.001, 0.001)
sigma0 <- sqrt(1/tau0)
tau1 ~ dgamma(0.001, 0.001)
sigma1 <- sqrt(1/tau1)
}
"
And run model
library(rjags)
mod <- jags.model( textConnection(st),
data=list(y=qq$yN,
x=qq$x,
ind=qq$indiv,
group=qq$group,
n=nrow(qq),
nInd=length(unique(qq$indiv)),
nGrp=length(unique(qq$group))),
n.adapt=1e6,
inits=list(.RNG.seed=1,
.RNG.name="base::Wichmann-Hill")
)
mod <- coda.samples(mod,
variable.names=c("beta","b1", "b2", "sigma", "sigma0", "sigma1"),
n.iter=1e6,
thin=5)
summary(mod)
qq <- structure(list(yN = c(3.51, 5.13, 5.2, 7.46, 5.64, 5.14, 6.84,
7.19, 7.77, 6, 10.97, 9.75, 5.43, 1.11, 10.31, 5.3, 4.52, 4.62,
3.97, 4.31, 8.2, 7.24, 6.75, 0, 7.77, 4.25, 5.29, 2.46, 4.3,
6.67, 8.72, 7.52, 6.12, 6.02, 1.48, 4.65, 7.52, 5.88, 6.06, 5.27,
6.04, 5.36, 7.34, 6.39, 2.84, 3.95, 8.07, 7.22, 4.78, 9.92, 5.85,
2.75, 6.34, 2.62, 7.3, 15.45, 5, 1.52, 8.3, 6.25, 16.32, 5.67,
8.55, 5.72, 2.8, 6.06, 1.3, 11.74, 7.02, 12.85, 6.46, 3.68, 8.48,
0.28, 0.92), x = c(-0.63, 0.18, -0.84, 1.6, 0.33, -0.82, 0.49,
0.74, 0.58, -0.31, 1.51, 0.39, -0.62, -2.21, 1.12, -0.04, -0.02,
0.94, 0.82, 0.59, 0.92, 0.78, 0.07, -1.99, 0.62, -0.06, -0.16,
-1.47, -0.48, 0.42, 1.36, -0.1, 0.39, -0.05, -1.38, -0.41, -0.39,
-0.06, 1.1, 0.76, -0.16, -0.25, 0.7, 0.56, -0.69, -0.71, 0.36,
0.77, -0.11, 0.88, 0.4, -0.61, 0.34, -1.13, 1.43, 1.98, -0.37,
-1.04, 0.57, -0.14, 2.4, -0.04, 0.69, 0.03, -0.74, 0.19, -1.8,
1.47, 0.15, 2.17, 0.48, -0.71, 0.61, -0.93, -1.25), indiv = structure(c(1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 4L, 4L,
4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L,
7L, 7L, 8L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 9L, 10L, 10L, 10L,
10L, 10L, 11L, 11L, 11L, 11L, 11L, 12L, 12L, 12L, 12L, 12L, 13L,
13L, 13L, 13L, 13L, 14L, 14L, 14L, 14L, 14L, 15L, 15L, 15L, 15L,
15L), .Label = c("a", "b", "c", "d", "e", "f", "g", "h", "i",
"j", "k", "l", "m", "n", "o"), class = "factor"), group = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L), .Label = c("A", "B",
"C", "D", "E"), class = "factor")), .Names = c("yN", "x", "indiv",
"group"), row.names = c(NA, -75L), class = "data.frame")
In a similar example, the nested structure of the data can be accounted for by creating an interaction variable, and using that as the grouping variable (so similar to the previous example of unique sets within groups).
data(Pastes, package="lme4")
lmer(strength ~ 1 + (1|batch/cask), data=Pastes)
lmer(strength ~ 1 + (1|batch) + (1|batch:cask), data=Pastes) # equivalent
How can this be coded in jags, and can it be done without creating an intermediate interaction variable?

beta[1]back to eachb1the estimated values should be very close to what is reported in lme4. – Onestepbpredictions should be the same asranef(lme4model)– Snowshedlmermodels at the start, there is no correlation between random terms defined or returned (if i understand correctly) – Snowshed