Pictures are better than words, so please have a look at 
What I have is
- a RasterLayer object (filled with random values here for illustration purposes only, the actual values don't matter)
- a SpatialPolygons object with lots and lots of polygons in it
You can re-create the example data I used for the image with the following code:
library(sp)
library(raster)
library(rgeos)
# create example raster
r <- raster(nrows=10, ncol=15, xmn=0, ymn=0)
values(r) <- sample(x=1:1000, size=150)
# create example (Spatial) Polygons
p1 <- Polygon(coords=matrix(c(50, 100, 100, 50, 50, 15, 15, 35, 35, 15), nrow=5, ncol=2), hole=FALSE)
p2 <- Polygon(coords=matrix(c(77, 123, 111, 77, 43, 57, 66, 43), nrow=4, ncol=2), hole=FALSE)
p3 <- Polygon(coords=matrix(c(110, 125, 125, 110, 67, 75, 80, 67), nrow=4, ncol=2), hole=FALSE)
lots.of.polygons <- SpatialPolygons(list(Polygons(list(p1, p2, p3), 1)))
crs(lots.of.polygons) <- crs(r) # copy crs from raster to polygons (please ignore any potential problems related to projections etc. for now)
# plot both
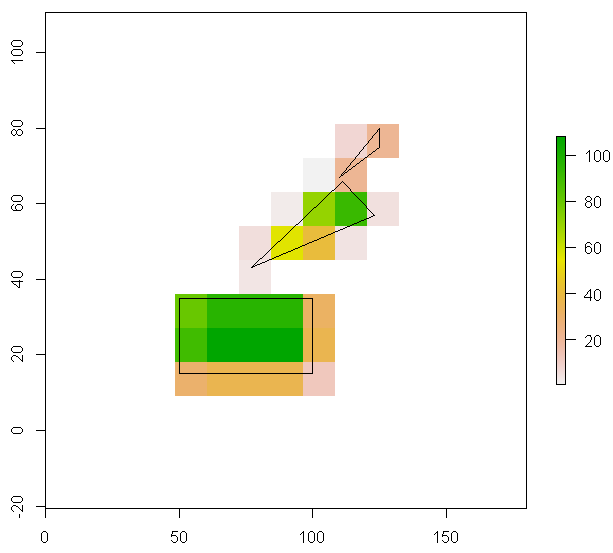
plot(r) #values in this raster for illustration purposes only
plot(lots.of.polygons, add=TRUE)
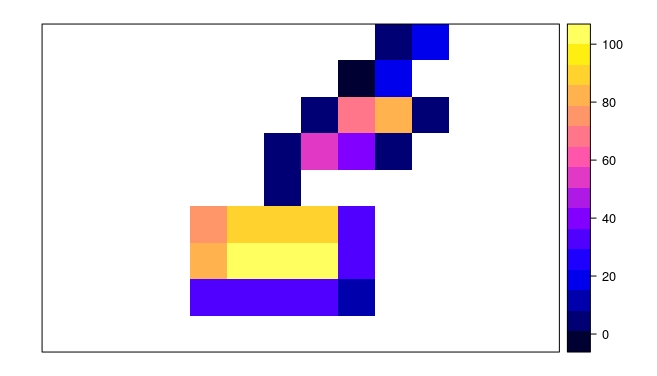
For each cell in the raster, I want to know how much of it is covered by one or more polygons. Or actually: the area of all polygons within the raster cell, without what is outside the cell in question. If there are multiple polygons overlapping a cell, I only need their combined area.
The following code does what I want, but takes more than a week to run with the actual data sets:
# empty the example raster (don't need the values):
values(r) <- NA
# copy of r that will hold the results
r.results <- r
for (i in 1:ncell(r)){
r.cell <- r # fresh copy of the empty raster
r.cell[i] <- 1 # set the ith cell to 1
p <- rasterToPolygons(r.cell) # create a polygon that represents the i-th raster cell
cropped.polygons <- gIntersection(p, lots.of.polygons) # intersection of i-th raster cell and all SpatialPolygons
if (is.null(cropped.polygons)) {
r.results[i] <- NA # if there's no polygon intersecting this raster cell, just return NA ...
} else{
r.results[i] <- gArea(cropped.polygons) # ... otherwise return the area
}
}
plot(r.results)
plot(lots.of.polygons, add=TRUE)
I can squeeze out a bit more speed by using sapply instead of the for-loop, but the bottleneck seems to be somewhere else. The whole approach feels pretty awkward, and I'm wondering if I've missed something obvious. At first I thought rasterize() should be able to do this easily, but I can't figure out what to put into the fun= argument. Any ideas?

foreachloops to use your multiple cores (if available). – Respectability