I am learning how to handle the FFTW package with fortran. To produce an easily verifiable example, I compute the power spectrum of a 2d plane, which I fill with two distinct superpositioned waves. This way, I know exactly where to expect peaks in the power spectrum.
As the FFTW documentation is more extensive for C, I first implemented the algorithm in C, which gives me quite satisfactory results:
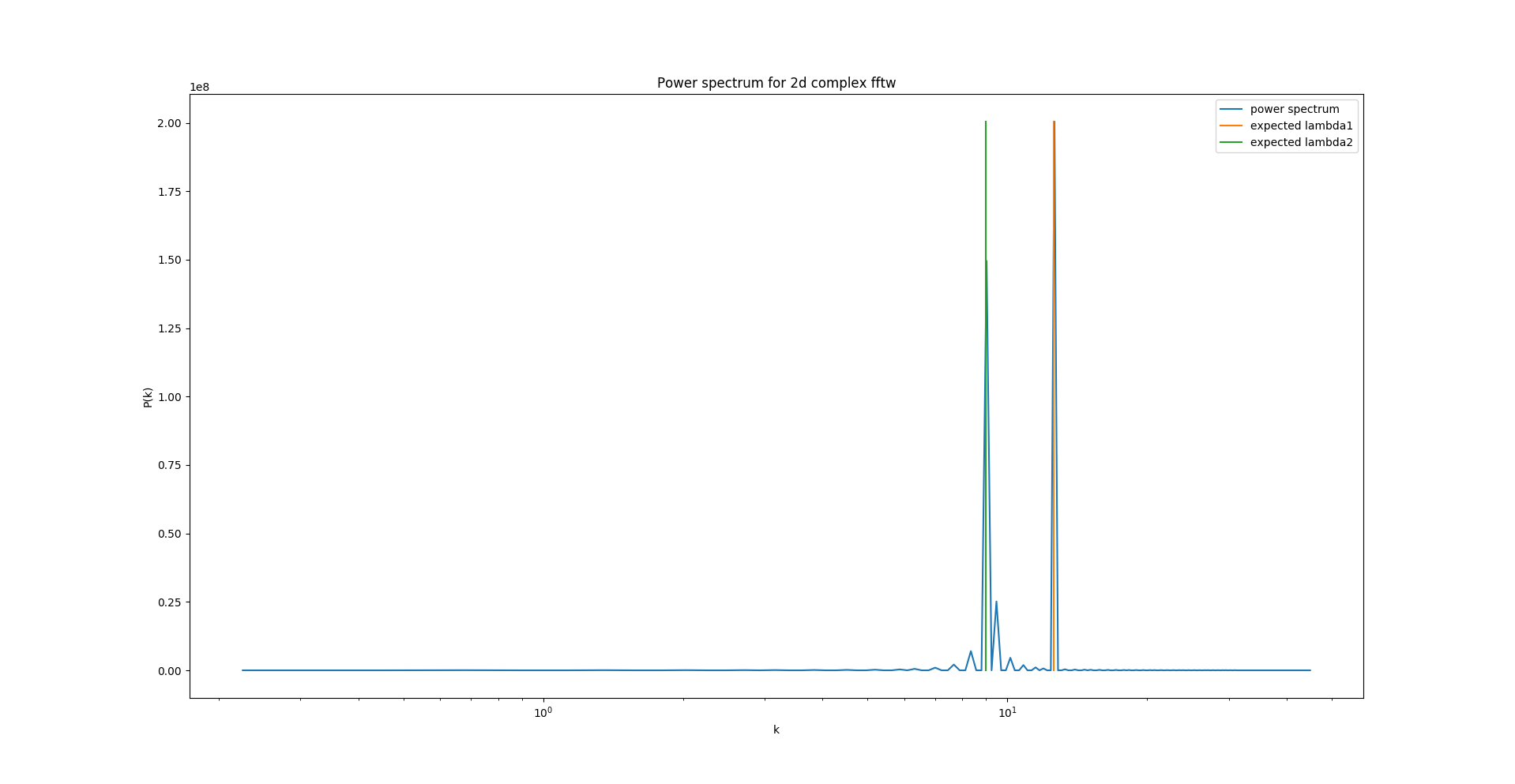 "lambda1" and "lambda2" are the known, expected wavelengths. The blue line is the obtained power spectrum.
"lambda1" and "lambda2" are the known, expected wavelengths. The blue line is the obtained power spectrum.
Then I tried to do exactly the same with fortran, which gives weird results:
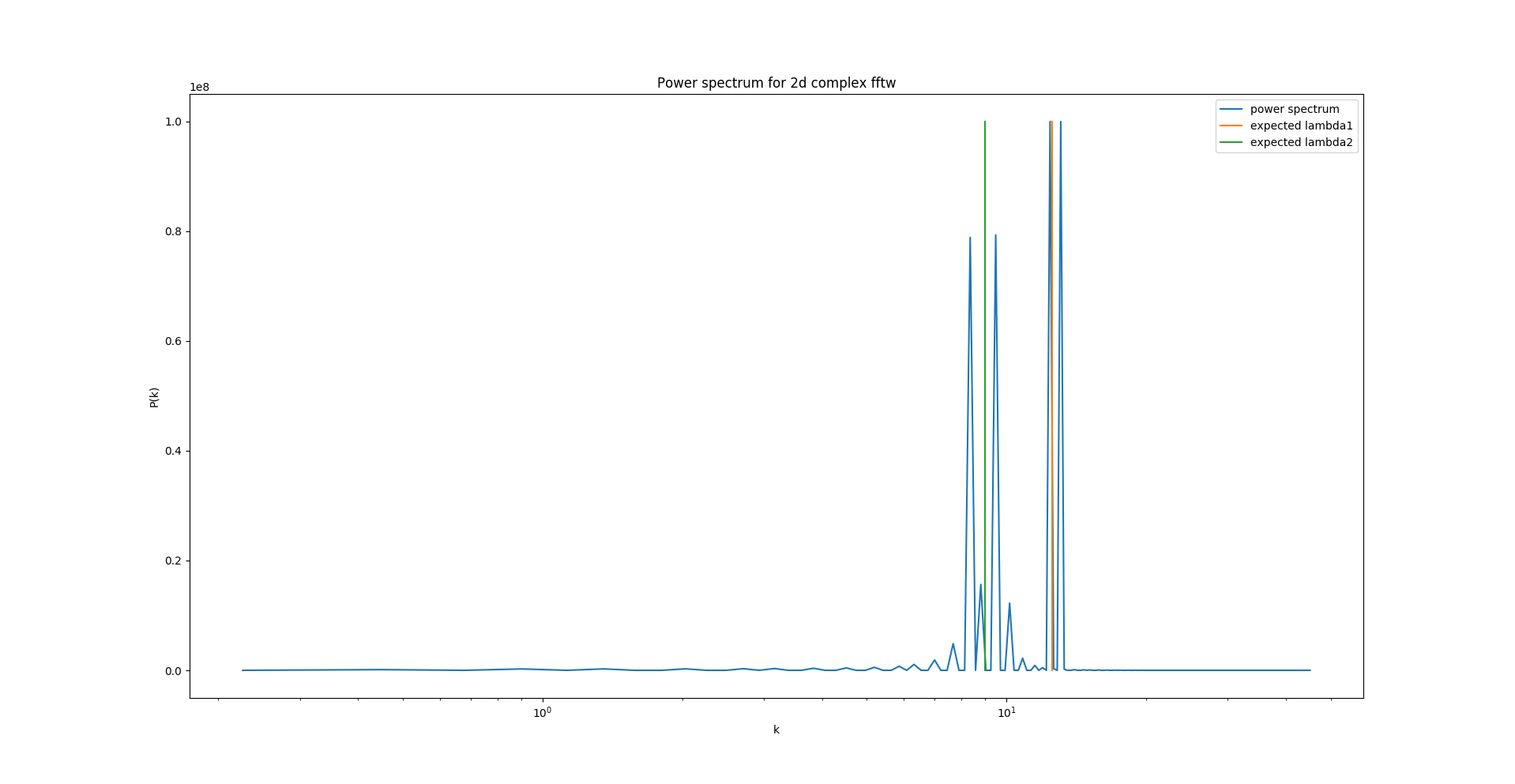
I have no idea where to look for possible errors. The codes execute without a hitch. Can anybody help?
Here is the C code, compiled with: gcc stackexchange.c -o a.out -I/home/myname/.local/include -lfftw3 -lm -g -Wall (gcc 5.4.0)
#include <fftw3.h>
#include <math.h>
#include <stdio.h>
#include <stdlib.h>
int main(void)
{
// parameters
int Nx = 200;
int Ny = 100;
int nsamples = 200;
float pi = 3.1415926;
float physical_length_x = 20;
float physical_length_y = 10;
float lambda1 = 0.5;
float lambda2 = 0.7;
float dx = physical_length_x/Nx;
float dy = physical_length_y/Ny;
float dkx = 2*pi/physical_length_x;
float dky = 2*pi/physical_length_y;
// counters/iterators
int ind, i, j, ix, iy, ik;
// power spectra stuff
float *Pk, *distances_k, d, kmax;
float *Pk_field;
// FFTW vars and arrays
fftw_complex *in, *out;
fftw_plan my_plan;
// allocate arrays for input/output
in = (fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx * Ny);
out = (fftw_complex *) fftw_malloc(sizeof(fftw_complex) * Nx * Ny);
// Create Plan
int n[2]; n[0] = Nx; n[1] = Ny;
my_plan = fftw_plan_dft(2, n, in, out, FFTW_FORWARD, FFTW_ESTIMATE);
// fill up array with a wave
for (i=0; i<Nx; i++){
for (j=0; j<Ny; j++){
ind = i*Ny + j;
in[ind][0] = cos(2.0*pi/lambda1*i*dx) + sin(2.0*pi/lambda2*j*dy);
}
}
// execute fft
fftw_execute(my_plan); /* repeat as needed */
// Calculate power spectrum: P(kx, ky) = |F[delta(x,y)]|^2
Pk_field = malloc(sizeof(float)*Nx*Ny);
for (i=0; i<Nx; i++){
for (j=0; j<Ny; j++){
ind = i*Ny+j;
Pk_field[ind] = out[ind][0]*out[ind][0] + out[ind][1]*out[ind][1];
}
}
Pk = calloc(nsamples, sizeof(float));
distances_k = malloc(nsamples*sizeof(float));
kmax = sqrt(pow((Nx/2+1)*dkx, 2) + pow((Ny/2+1)*dky, 2));
for(i=0; i<nsamples; i++){
distances_k[i]= 1.0001*i/nsamples*kmax; // add a little more to make sure kmax will fit
}
// histogrammize P(|k|)
for (i=0; i<Nx; i++){
if (i<Nx/2+1)
{ ix = i; }
else
{ ix = -Nx+i; }
for (j=0; j<Ny; j++){
if (j<Ny/2+1)
{ iy = j; }
else
{ iy = -Ny+j; }
ind = i*Ny + j;
d = sqrt(pow(ix*dkx,2)+pow(iy*dky,2));
for(ik=0; ik<nsamples; ik++){
if (d<=distances_k[ik] || ik==nsamples){
break;
}
}
Pk[ik] += Pk_field[ind];
}
}
//-----------------------------------
// write arrays to file.
// can plot them with plot_fftw.py
//-----------------------------------
FILE *filep;
filep = fopen("./fftw_output_2d_complex.txt", "w");
for (i=0; i<nsamples; i++){
fprintf(filep, "%f\t%f\n", distances_k[i], Pk[i]);
}
fclose(filep);
printf("Finished! Written results to ./fftw_output_2d_complex.txt\n");
//----------------------
// deallocate arrays
//----------------------
fftw_destroy_plan(my_plan);
fftw_free(in); fftw_free(out);
return 0;
}
This is the Fortran code, compiled with: gfortran stackexchange.f03 -o a.out -L/home/my_name/.local/lib/ -I/home/my_name/.local/include/ -lfftw3 -g -Wall
program use_fftw
use,intrinsic :: iso_c_binding
implicit none
include 'fftw3.f03'
integer, parameter :: dp=kind(1.0d0)
integer, parameter :: Nx = 200
integer, parameter :: Ny = 100
integer, parameter :: nsamples = 200
real(dp), parameter :: pi = 3.1415926d0
real(dp), parameter :: physical_length_x = 20.d0
real(dp), parameter :: physical_length_y = 10.d0
real(dp), parameter :: lambda1 = 0.5d0
real(dp), parameter :: lambda2 = 0.7d0
real(dp), parameter :: dx = physical_length_x/real(Nx,dp)
real(dp), parameter :: dy = physical_length_y/real(Ny,dp)
real(dp), parameter :: dkx = 2.d0 * pi / physical_length_x
real(dp), parameter :: dky = 2.d0 * pi / physical_length_y
integer :: i, j, ix, iy, ik
real(dp):: kmax, d
complex(dp), allocatable, dimension(:,:) :: arr_in, arr_out, Pk_field
real(dp), allocatable, dimension(:) :: Pk, distances_k
integer*8 :: my_plan
integer :: n(2) = (/Nx, Ny/)
allocate(arr_in( 1:Nx,1:Ny))
allocate(arr_out(1:Nx,1:Ny))
! call dfftw_plan_dft_2d(my_plan, Nx, Ny, arr_in, arr_out, FFTW_FORWARD, FFTW_ESTIMATE) ! doesn't help either
call dfftw_plan_dft(my_plan, 2, n, arr_in, arr_out, FFTW_FORWARD, FFTW_ESTIMATE)
! fill up wave
do i = 1, Nx
do j = 1, Ny
arr_in(i,j) =cmplx(cos(2.0*pi/lambda1*i*dx)+sin(2.0*pi/lambda2*j*dy) , 0.d0, kind=dp)
enddo
enddo
! execute fft
call dfftw_execute_dft(my_plan, arr_in, arr_out)
allocate(Pk_field(1:Nx, 1:Ny))
allocate(distances_k(1:nsamples))
allocate(Pk(1:nsamples))
Pk=0
distances_k=0
! Get bin distances
kmax = sqrt(((Nx/2+1)*dkx)**2+((Ny/2+1)*dky)**2)*1.001
do i = 1, nsamples
distances_k(i) = i*kmax/nsamples
enddo
! Compute P(k) field, distances field
do i = 1, Nx
do j = 1, Ny
Pk_field(i,j) = arr_out(i,j)*conjg(arr_out(i,j))
if (i<=Nx/2+1) then
ix = i
else
ix = -Nx+i
endif
if (j<=Ny/2+1) then
iy = j
else
iy = -Ny+j
endif
d = sqrt((ix*dkx)**2+(iy*dky)**2)
do ik=1, nsamples
if (d<=distances_k(ik) .or. ik==nsamples) exit
enddo
Pk(ik) = Pk(ik)+real(Pk_field(i,j))
enddo
enddo
! write file
open(unit=666,file='./fftw_output_2d_complex.txt', form='formatted')
do i = 1, nsamples
write(666, '(2E14.5,x)') distances_k(i), Pk(i)
enddo
close(666)
deallocate(arr_in, arr_out, Pk, Pk_field, distances_k)
call dfftw_destroy_plan(my_plan)
write(*,*) "Finished! Written results to fftw_output_2d_complex.txt"
end program use_fftw
For your convenience, this is my short python script that I use to plot the results:
#!/usr/bin/python3
#====================================
# Plots the results of the FFTW
# example programs.
#====================================
import numpy as np
import matplotlib.pyplot as plt
from sys import argv
from time import sleep
errormessage="""
I require an argument: Which output file to plot.
Usage: ./plot_fftw.py <case>
options for case:
1 fftw_1d_complex.txt
2 fftw_2d_complex.txt
3 fftw_1d_real.txt
4 fftw_2d_real.txt
Please select a case: """
#----------------------
# Hardcoded stuff
#----------------------
file_dict={}
file_dict['1'] = ('fftw_output_1d_complex.txt', '1d complex fftw')
file_dict['2'] = ('fftw_output_2d_complex.txt', '2d complex fftw')
file_dict['3'] = ('fftw_output_1d_real.txt', '1d real fftw')
file_dict['4'] = ('fftw_output_2d_real.txt', '2d real fftw')
lambda1=0.5
lambda2=0.7
#------------------------
# Get case from cmdline
#------------------------
case = ''
def enforce_integer():
global case
while True:
case = input(errormessage)
try:
int(case)
break
except ValueError:
print("\n\n!!! Error: Case must be an integer !!!\n\n")
sleep(2)
if len(argv) != 2:
enforce_integer()
else:
try:
int(argv[1])
case = argv[1]
except ValueError:
enforce_integer()
filename,title=file_dict[case]
#-------------------------------
# Read and plot data
#-------------------------------
k, Pk = np.loadtxt(filename, dtype=float, unpack=True)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.set_title("Power spectrum for "+title)
ax.set_xlabel("k")
ax.set_ylabel("P(k)")
# ax.plot(k, Pk, label='power spectrum')
ax.semilogx(k[k>0], Pk[k>0], label='power spectrum') # ignore negative k
ax.plot([2*np.pi/lambda1]*2, [Pk.min()-1, Pk.max()+1], label='expected lambda1')
ax.plot([2*np.pi/lambda2]*2, [Pk.min()-1, Pk.max()+1], label='expected lambda2')
ax.legend()
plt.show()

0,1offset thing when computing/comparingdistances_k(still guessing)? – Autrey