My question is related to this one. I have a file named 'test.csv' with 'NA' as a value for region. I want to read this in as 'NA', not 'NaN'. However, there are missing values in other columns in test.csv, which I want to retain as 'NaN'. How can I do this?
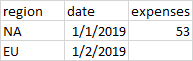
# test.csv looks like this:
Here's what I've tried:
import pandas as pd
# This reads NA as NaN
df = pd.read_csv(test.csv)
df
region date expenses
0 NaN 1/1/2019 53
1 EU 1/2/2019 NaN
# This reads NA as NA, but doesn't read missing expense as NaN
df = pd.read_csv('test.csv', keep_default_na=False, na_values='_')
df
region date expenses
0 NA 1/1/2019 53
1 EU 1/2/2019
# What I want:
region date expenses
0 NA 1/1/2019 53
1 EU 1/2/2019 NaN
The problem with adding the argument keep_default_na=False is that the second value for expenses does not get read in as NaN. So if I then try pd.isnull(df['value'][1]) this is returned as False.

nullvalues are represented by an underscore hence they setna_values='_'. In your case missing data appear to be represented by the empty string, so I'd go withna_values=''(in addition tokeep_default_na=False) If that solves your problem then this is clearly a dup. – Harping