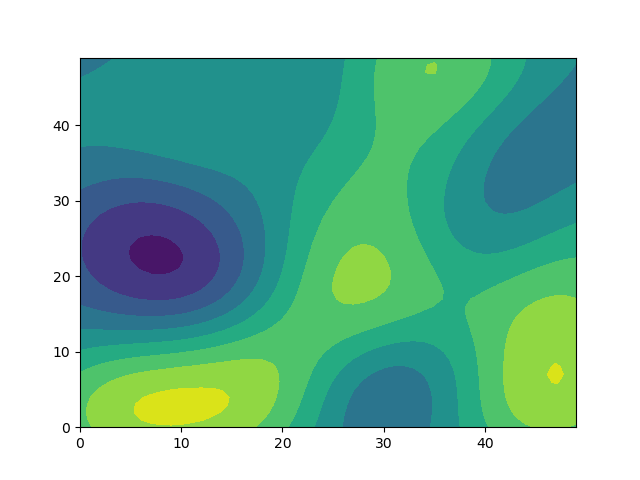
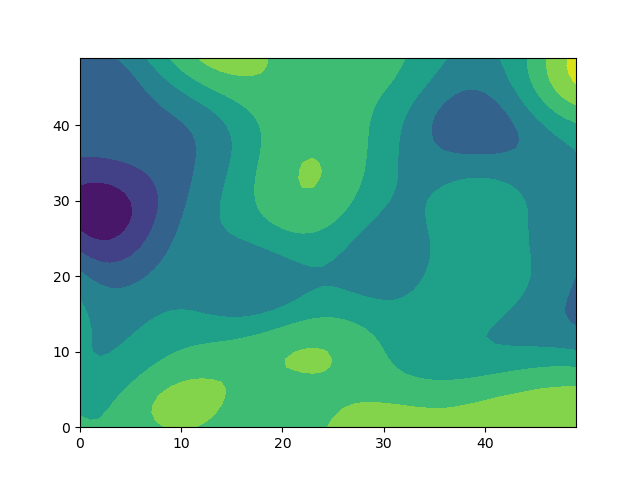
In my current project, I am interested in calculating spatially correlated noise for a large model grid. The noise should be strongly correlated over short distances, and uncorrelated over large distances. My current approach uses multivariate Gaussians with a covariance matrix specifying the correlation between all cells.
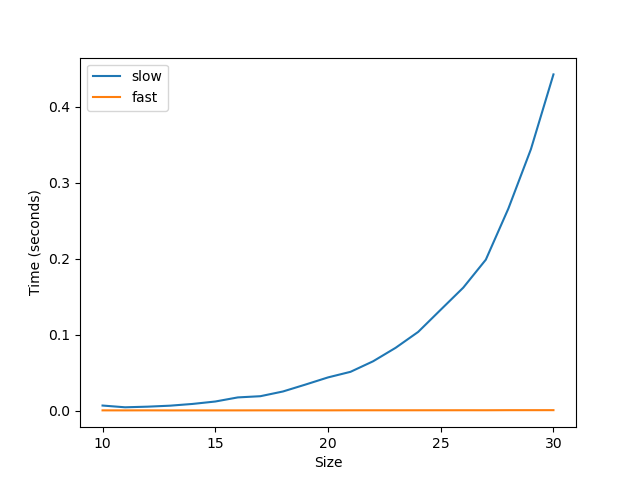
Unfortunately, this approach is extremely slow for large grids. Do you have a recommendation of how one might generate spatially correlated noise more efficiently? (It doesn't have to be Gaussian)
import scipy.stats
import numpy as np
import scipy.spatial.distance
import matplotlib.pyplot as plt
# Create a 50-by-50 grid; My actual grid will be a LOT larger
X,Y = np.meshgrid(np.arange(50),np.arange(50))
# Create a vector of cells
XY = np.column_stack((np.ndarray.flatten(X),np.ndarray.flatten(Y)))
# Calculate a matrix of distances between the cells
dist = scipy.spatial.distance.pdist(XY)
dist = scipy.spatial.distance.squareform(dist)
# Convert the distance matrix into a covariance matrix
correlation_scale = 50
cov = np.exp(-dist**2/(2*correlation_scale)) # This will do as a covariance matrix
# Sample some noise !slow!
noise = scipy.stats.multivariate_normal.rvs(
mean = np.zeros(50**2),
cov = cov)
# Plot the result
plt.contourf(X,Y,noise.reshape((50,50)))