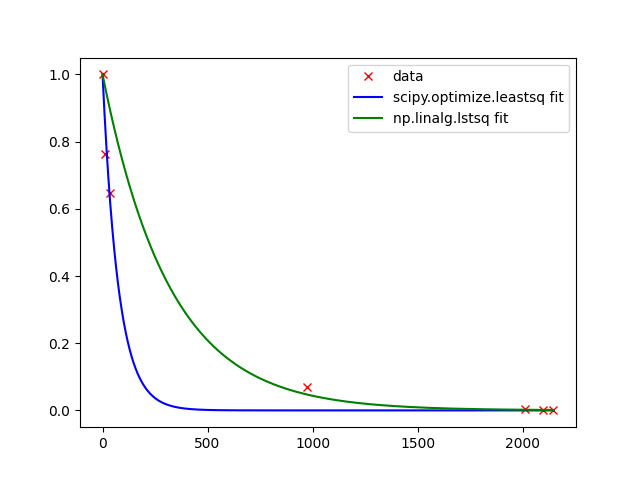
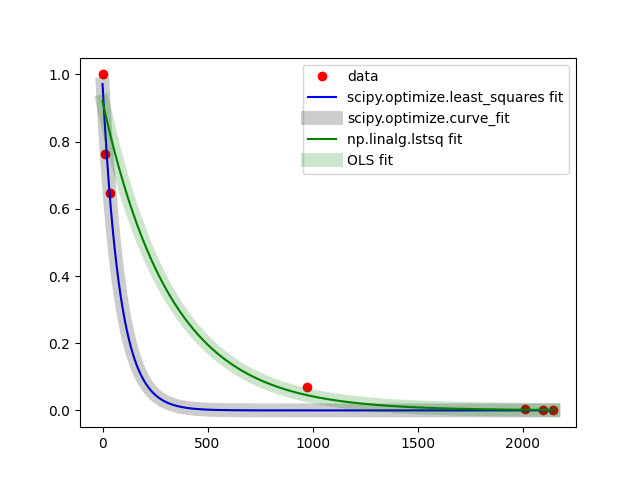
of course, possibility to linearize data (your case) and use of LS-approach log_transformed_fit here is always faster and for full-rank and well-conditioned(not singular) matrices gives best fit (as it is BLUE). Linear Algebra is always comparably fast in numpy and scipy. Non-linear approximation using finite-differences is the last choice from speed viewpoint. If you can provide explicitly jac and hess functions for non-linear approximation, of course you will benefit for speed. Using non-linear approximation with many of scipy.optimize methods you always have threat to stack in local minimum under certain chosen init_guess_params -- therefore only convex functions with one optimum value can give correct resulted parameters. Otherwise, you can just choose more suitable method from those available in scipy.optimize-package. For global min/max in scipy.optimize-package other functions exist. So everything, as usually, depends on Data, Task and Solver. For the use of scipy.optimize.least_squares can try to vary loss and f_scale depending on data. Of course, statistical scores (e.g. goodness-of-fit etc) can help to choose the best solver (for best fit). But again, OLS gives BLUE estimates and for the predictive ability model should be efficient and consistent as well, you have small size of sample. For your data linear and non-linear fitting looks like this:
import scipy.optimize as optimize
import numpy as np
import matplotlib.pyplot as plt
## Sampled data
x = np.array([0, 14, 37, 975, 2013, 2095, 2147])
y = np.array([1.0, 0.764317544, 0.647136491, 0.070803763, 0.003630962, 0.001485394, 0.000495131])
x_ = np.arange(x.min(), x.max(), 0.2) # grid for plot
plt.figure()
plt.plot(x, y, 'ro', label= 'data')
################################### Normal Equation
##a = np.vstack([x, np.ones(len(x))]).T
##print(np.dot(np.linalg.inv(np.dot(a.T, a)), np.dot(a.T, y)))
###################################
## scipy.optimize FOR NON-LINEAR APPROX.
################################### scipy.optimize.least_squares
# function
fp = lambda a, p, x: a * np.exp(p*x)
# error function
e = lambda p, x, y: (y - fp(*p, x) )
# using scipy least squares
res = optimize.least_squares(e, [0., 0.], args=(x,y), )
##print( res)
l1 = res.x
print( l1)
plt.plot(x_, fp(*l1, x_), 'b-', label= 'scipy.optimize.least_squares fit')
##full_output
##popt, pcov, info, mesg, ler = optimize.leastsq(residual, p0, args=(x, y), full_output=True)
##print("popt, pcov, info, mesg, ler: ", popt, pcov, info, mesg, ler)
####################################### scipy.optimize.curve_fit
##if p0=(1, 1) then
##RuntimeWarning: overflow encountered in exp: return a*np.exp(-c*x)
from scipy.optimize import curve_fit
def func(x, a, c):
return a*np.exp(c*x)
popt, pcov = curve_fit(func, x, y, p0=(1, 0))
print(popt)
y_cf = func(x_, *popt)
plt.plot(x_, y_cf, 'k-', linewidth= 10, label= 'scipy.optimize.curve_fit', alpha=0.2 )
####################################
## LS FOR LINEAR PARAMS
#################################### np.linalg.lstsq
# using numpy least squares
xr, residuals, rank, s = np.linalg.lstsq(np.vstack([x, np.ones(len(x))]).T, np.log(y), rcond=-1)
##print("x, residuals, rank, s: ", x, residuals, rank, s)
l2= xr
print( "slope, intercept: ", l2)
# .......... (same answer as Excel trend line)
y_2 = np.exp(l2[1]) * np.exp(l2[0] * x_)
plt.plot(x_, y_2, 'g-', label= 'np.linalg.lstsq fit')
#################################### statsmodels
import statsmodels.api as sm
X = sm.add_constant(x)
model = sm.OLS(np.log(y), X)
result = model.fit()
intercept_OLS, slope_OLS = result.params
print('OLS: ', slope_OLS, intercept_OLS,)
y_3 = np.exp(intercept_OLS) * np.exp( slope_OLS * x_)
plt.plot(x_, y_3, 'g-', label= 'OLS fit', linewidth= 10, alpha=0.2)
plt.legend()
plt.show()
![enter image description here]()
with data from the example from docs plot will be significantly different
Normalizing data and then Linearizing y=Ae^(Bx) taking logarithm from both sides lny=lnA+Bx gives the possibility to use least-squares from scipy.optimize
xxx= x
yyy= np.log(y)
# model function
model = lambda a, b, x: (np.log(a) + b*x)
# error function
def e ( p, x, y): return (y - model(*p, x) ) #**2
# using scipy least squares
params = optimize.least_squares(e, [1., 0.], args=(xxx,yyy), ).x
plt.plot(x_, np.exp(model(*params, x_)), 'm-', label= ' scipy.optimize.least_squares for linearized data'); plt.legend()
p.s. other linearization techniques