TL;DR: Set ticks and ticklabels to different values
When extent= is set to some list, the image is stretched individually along x- and y-axes to fill the box. But sometimes, it's still better to set the tick labels explicitly (imo) using ax.set or plt.xticks/plt.yticks:
fig, ax = plt.subplots(figsize=(6,6))
ax.imshow(hist, cmap='Reds', interpolation='none', extent=[80, 120, 32, 0], aspect=2)
ax.set(xticks=np.arange(80, 122)[::10], xticklabels=np.arange(80, 122)[::10]);
Since extent= sets the image size, using it to set tick labels is sometimes not ideal. For example, say, we want to display an image that is long relatively tall but with small tick labels, such as the following:
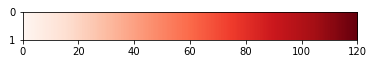
![correct output]()
Then,
fig, ax = plt.subplots(1, figsize=(6, 6))
ax.imshow(np.arange(120)[None, :], cmap='Reds', extent=[0, 120, 1, 0]);
produces
![wrong output]()
but
fig, ax = plt.subplots(1, figsize=(6, 6))
ax.imshow(np.arange(120)[None, :], cmap='Reds', extent=[0, 120, 10, 0]);
ax.set(xticks=np.linspace(0, 120, 7), xticklabels=np.arange(0, 121, 20), yticks=[0, 10], yticklabels=[0, 1]);
produces the correct output. That's because extent= was set to large values but the tick labels where set to smaller values so that the image has the desired labels.
N.B. ax.get_xticks() and ax.get_yticks() are useful methods to understand the default (or otherwise) tick locations and ax.get_xlim() and ax.get_ylim() are useful methods to understand the axes limits.
Even in the method used by OP, without any extent=, ax.get_xlim() returns (-1.0, 19.5). Since the x-tick location range is already set as such, it could be used to set x-tick labels to something else; simply set xticks to be some values within this range and assign whatever values to xticklabels. So the following renders the desired image.
fig, ax = plt.subplots(figsize=(6,6))
ax.imshow(hist, cmap='Reds', interpolation='none', aspect=2)
ax.set(xticks=np.arange(-1, 20, 5), xticklabels=np.arange(80, 122, 10));





pcolorinstead ofimshowas mentioned in this answer. – Fagot