I am doing a multiple regression in Stan.
I want a trace plot of the beta vector parameter for the regressors/design matrix.
When I do the following:
fit = model.sampling(data=data, iter=2000, chains=4)
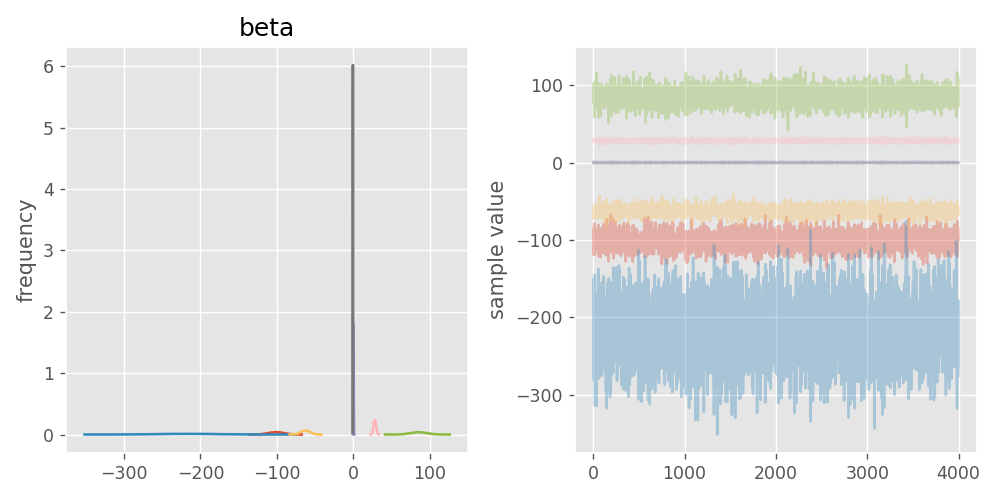
fig = fit.plot('beta')
I get a pretty horrid image:
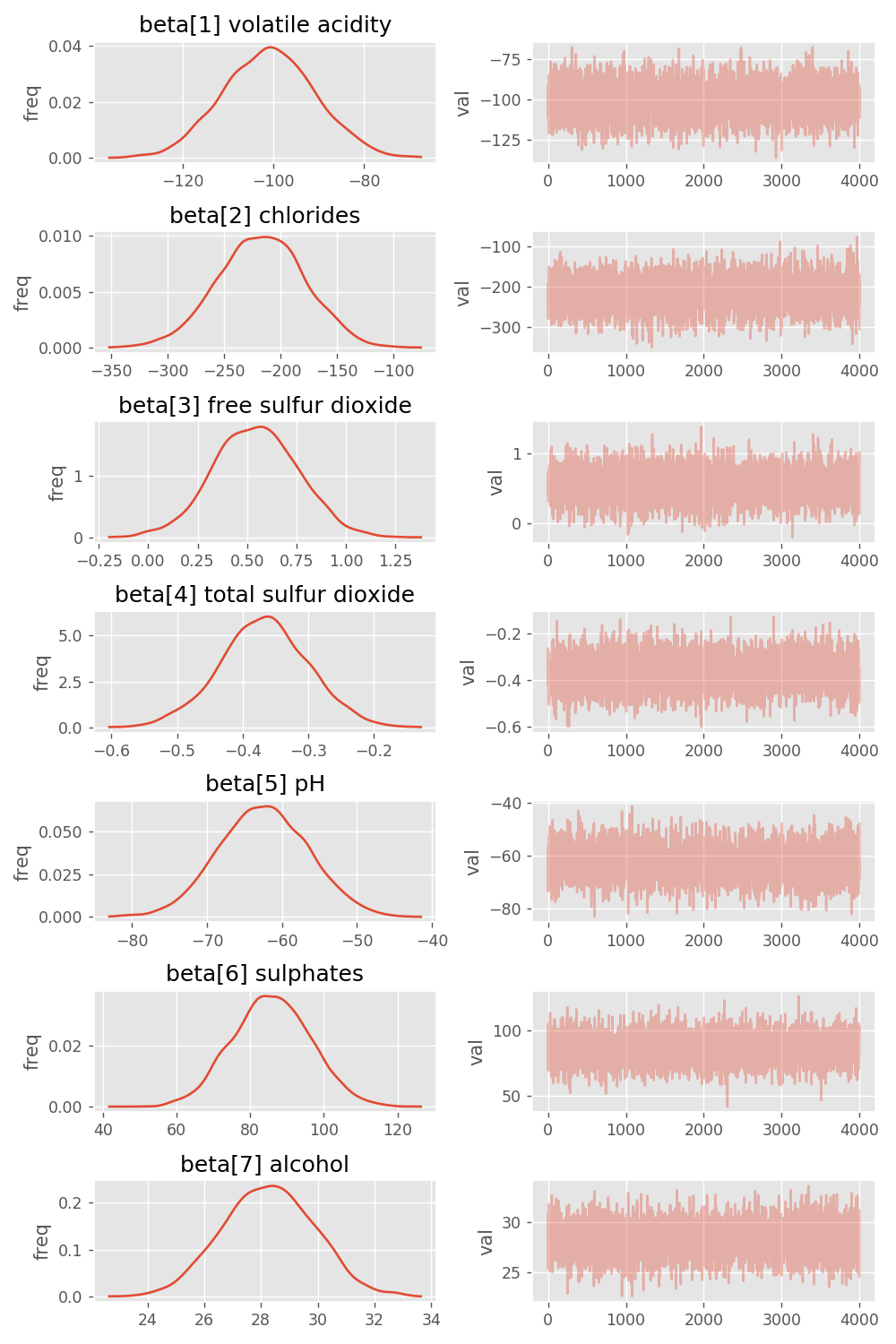
I was after something a little more user friendly. I have managed to hack the following which is closer to what I am after.
My hack plugs into the back of pystan as follows.
r = fit.extract() # r for results
from pystan.external.pymc import plots
param = 'beta'
beta = r[param]
name = df.columns.values.tolist()
(rows, cols) = beta.shape
assert(len(df.columns) == cols)
values = {param+'['+str(k+1)+'] '+name[k]:
beta[:,k] for k in range(cols)}
fig = plots.traceplot(values, values.keys())
for a in fig.axes:
# shorten the y-labels
l = a.get_ylabel()
if l == 'frequency':
a.set_ylabel('freq')
if l=='sample value':
a.set_ylabel('val')
fig.set_size_inches(8, 12)
fig.tight_layout(pad=1)
fig.savefig(g_dir+param+'-trace.png', dpi=125)
plt.close()
My question - surely I have missed something - but is there an easier way to get the kind of output I am after from pystan for a vector parameter?