I did not find an existing solution for this, so I wrote a function which does what you need. Of course, it will give inappropriate results for big datasets.
require(dplyr)
timeline_plot <- function(dat, spacing = 0.01, team_size = 0.25, notch = 0.1,
cols = list(team = "lightblue",
completed = "green3",
"to do" = "lightgray"),
cex_label = 2){
# Arguments:
# dat = data frame
# spacing = space between polygons (part of plot width)
# team_size = size of team polygon (part of plot width)
# notch = size of arrow side protruding (part of plot width)
# cols = color for each status
# cex_lab = cex of labels
# Count number of columns
dat_n <- dat %>%
group_by(team) %>%
summarise(n = length(team))
# Get number of rows
nr <- length(dat_n$team)
# Prepare polygon
poly <- matrix(c(0, 0, 0, 1, 1, 1, 0, 0.5, 1, 1, 0.5, 0), ncol = 2)
# Function for polygon scaling, shifting and notch adding
morph_poly <- function(poly, scale_x = 1, shift_x = 0, notch){
poly[, 1] <- poly[, 1] * scale_x + shift_x
poly[c(2, 5), 1] <- poly[c(2, 5), 1] + notch
return(poly)
}
# Fucntion for label positioning
label_pos_x <- function(poly){
x <- poly[2, 1] + (poly[5, 1] - poly[2, 1]) / 3
return(x)
}
# Save old par
opar <- par()
# Set number of rows for plotting
par(mfrow = c(nr, 1))
par(mar = c(0,0,0,0))
# Actual plotting
for (i in c(1:nr)){
# Each row will be presentd as
# team_polygon + spacing + n * (spacing + task_polygon) + notch
team <- dat_n$team[i]
tasks <- dat[dat$team == team, ]
tasks <- tasks[order(tasks$task), ]
# Create empty plot
plot(NA, xlim = c(0, 1), ylim = c(0, 1), xlab = "", ylab = "", bty = "n", xaxt = "n", yaxt = "n")
# Plot team polygon
team_poly <- morph_poly(poly, team_size, 0, notch)
polygon(team_poly, col = cols$team)
# Add team label
text(label_pos_x(team_poly), 0.5, labels = dat_n$team[i], cex = cex_label)
# Calculate the size of task polygon
tasks_n <- dat_n$n[i]
size_x <- (1 - team_size - (tasks_n * spacing) - notch) / tasks_n
shift <- team_size + spacing
# plot each task polygon
for (j in 1:nrow(tasks)){
# Get task color
task_col = cols[[tasks$status[j]]]
# Prepare polygon
task_poly <- morph_poly(poly, scale_x = size_x, shift_x = shift + spacing, notch = notch)
polygon(task_poly, col = task_col)
# Add task label
text(label_pos_x(task_poly), 0.5, labels = tasks$task[j], cex = cex_label)
# Update shift
shift <- shift + size_x + spacing
}
}
# Set initial par
par(opar)
}
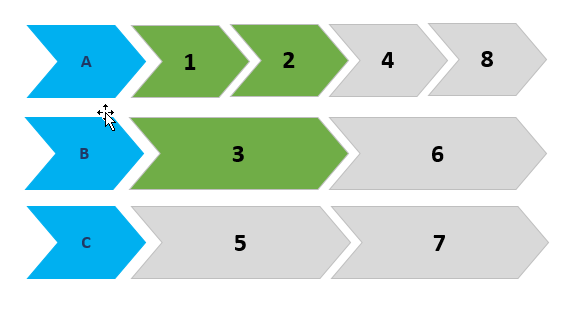
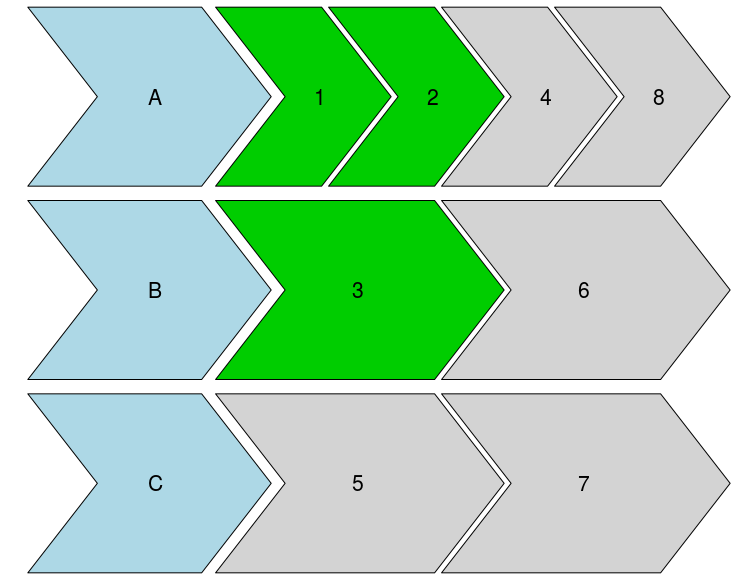
With your data set as dat it gives:
timeline_plot(dat)
![Function output]()

dput()to present your example data! – ScabbyDiagrammeRas stated. I don't think they have the shapes in the post though. – Geiger