I have data with two time axes and measurements for each cell. From this I create a heatmap. I also know for each cell whether the measurement is significant.
My problem is to draw a contour line around all cells that are significant. If cells form clusters with the same significance value, I need to draw the contour around the cluster and not around each individual cell.
The data are in this format:
x_time y_time metric signif
1 1 1 0.3422285 FALSE
2 2 1 0.6114085 FALSE
3 3 1 0.5381621 FALSE
4 4 1 0.5175120 FALSE
5 1 2 0.6997991 FALSE
6 2 2 0.3054885 FALSE
7 3 2 0.8353888 TRUE
8 4 2 0.3991566 TRUE
9 1 3 0.7522728 TRUE
10 2 3 0.5311418 TRUE
11 3 3 0.4972816 TRUE
12 4 3 0.4330033 TRUE
13 1 4 0.5157972 TRUE
14 2 4 0.6324151 TRUE
15 3 4 0.4734126 TRUE
16 4 4 0.4315119 TRUE
The code below generates this data, where the measurements are random (dt$metrics) and the significance is logical (dt$signif).
# data example
dt <- data.frame(x_time=rep(seq(1, 4), 4),
y_time=rep(seq(1, 4), each=4),
metric=(rnorm(16, 0.5, 0.2)),
signif=c(rep(FALSE, 6), rep(TRUE, 10)))
The heatmap alone can be generated using ggplot2's geom_tile
# Generate heatmap using ggplot2's geom_tile
library(ggplot2)
p <- ggplot(data = dt, aes(x = x_time, y = y_time))
p <- p + geom_tile(aes(fill = metric))
Based on this question, I managed to draw contours with different colors around each cell according to the significance value.
# Heatmap with lines around each significant cell
p <- ggplot(data = dt, aes(x = x_time, y = y_time))
p <- p + geom_tile(aes(fill = metric, color = signif), size = 2)
p <- p + scale_color_manual(values = c("black", "white"))

However, this approach does not group adjacent significant cells together by drawing a contour around the entire group (as is also discussed in the question I linked to).
As this question shows, it is possible to draw boxes around specified areas, but I do not think this can be extended to all possible clusters of cells.

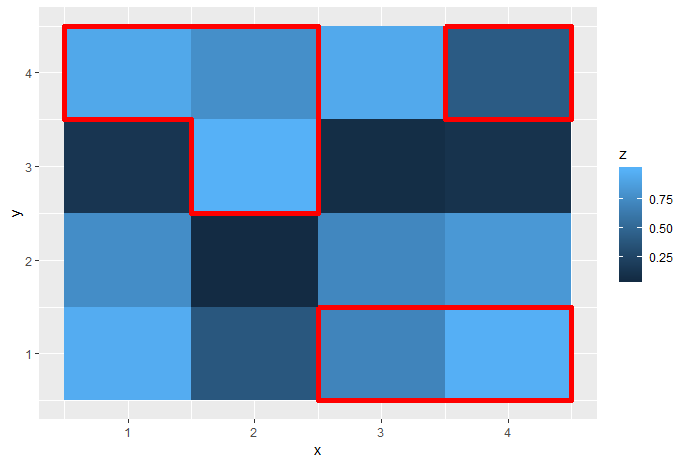
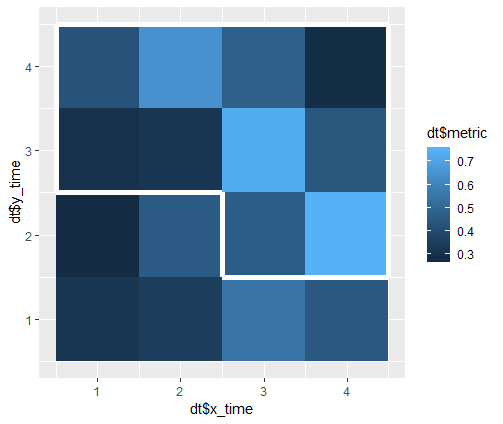
raster::clump. See e.g. How to get contour lines around the grids in R-raster? – Anceline