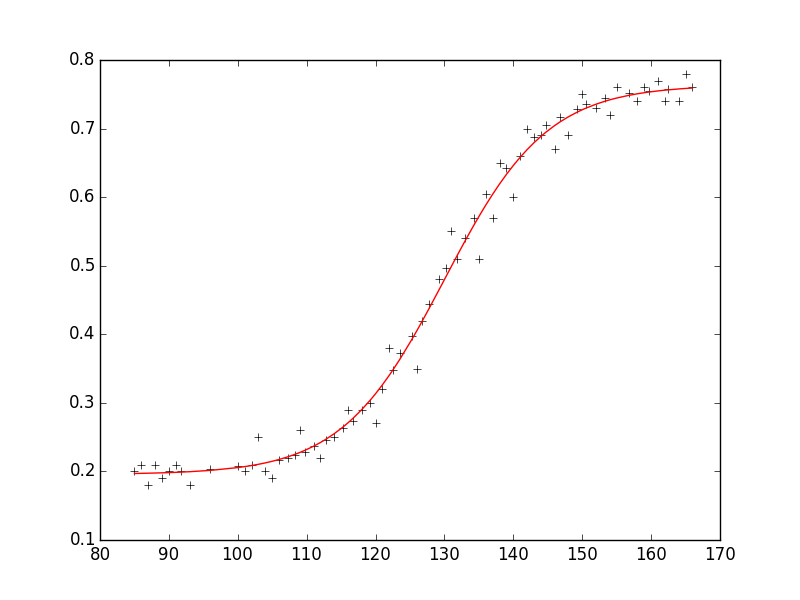
I have experimental data :
xdata = [85,86,87,88,89,90,91,91.75,93,96,100,101,102,103,104,105,106,107.25,108.25,109,109.75,111,112,112.75,114,115.25,116,116.75,118,119.25,120,121,122,122.5,123.5,125.25,126,126.75,127.75,129.25,130.25,131,131.75,133,134.25,135,136,137,138,139,140,141,142,143,144,144.75,146,146.75,148,149.25,150,150.5,152,153.25,154,155,156.75,158,159,159.75,161,162,162.5,164,165,166]
ydata = [0.2,0.21,0.18,0.21,0.19,0.2,0.21,0.2,0.18,0.204,0.208,0.2,0.21,0.25,0.2,0.19,0.216,0.22,0.224,0.26,0.229,0.237,0.22,0.246,0.25,0.264,0.29,0.274,0.29,0.3,0.27,0.32,0.38,0.348,0.372,0.398,0.35,0.42,0.444,0.48,0.496,0.55,0.51,0.54,0.57,0.51,0.605,0.57,0.65,0.642,0.6,0.66,0.7,0.688,0.69,0.705,0.67,0.717,0.69,0.728,0.75,0.736,0.73,0.744,0.72,0.76,0.752,0.74,0.76,0.7546,0.77,0.74,0.758,0.74,0.78,0.76]
And the formula f(x) = m1 + m2 / (1 + e ^ (-m3*(x - m4))). I need to find m1,
m2, m3, m4 with least square method, where
0.05 < m1 < 0.3
0.3 < m2 < 0.8
0.05 < m3 < 0.5
100 < m4 < 200.
I use curve_fit and my function is :
def f(xdata, m1, m2, m3, m4):
if m1 > 0.05 and m1 < 0.3 and \
m2 > 0.3 and m2 < 0.8 and \
m3 > 0.05 and m3 < 0.5 and \
m4 > 100 and m4 < 200:
return m1 + (m2 * 1. / (1 + e ** (-m3 * (x - m4))))
return (abs(m1) + abs(m2) + abs(m3) + abs(m4)) * 1e14 # some large number
But the program return the error: RuntimeError: Optimal parameters not found: Number of calls to function has reached maxfev = 1000.
What to do?
import numpy as np
from scipy.optimize import curve_fit
from math import e
xdata = np.array([85,86,87,88,89,90,91,91.75,93,96,100,101,102,103,104,105,106,107.25,108.25,109,109.75,111,112,112.75,114,115.25,116,116.75,118,119.25,120,121,122,122.5,123.5,125.25,126,126.75,127.75,129.25,130.25,131,131.75,133,134.25,135,136,137,138,139,140,141,142,143,144,144.75,146,146.75,148,149.25,150,150.5,152,153.25,154,155,156.75,158,159,159.75,161,162,162.5,164,165,166])`
ydata = np.array([0.2,0.21,0.18,0.21,0.19,0.2,0.21,0.2,0.18,0.204,0.208,0.2,0.21,0.25,0.2,0.19,0.216,0.22,0.224,0.26,0.229,0.237,0.22,0.246,0.25,0.264,0.29,0.274,0.29,0.3,0.27,0.32,0.38,0.348,0.372,0.398,0.35,0.42,0.444,0.48,0.496,0.55,0.51,0.54,0.57,0.51,0.605,0.57,0.65,0.642,0.6,0.66,0.7,0.688,0.69,0.705,0.67,0.717,0.69,0.728,0.75,0.736,0.73,0.744,0.72,0.76,0.752,0.74,0.76,0.7546,0.77,0.74,0.758,0.74,0.78,0.76])
def f(xdata, m1, m2, m3, m4):
if m1 > 0.05 and m1 < 0.3 and \
m2 > 0.3 and m2 < 0.8 and \
m3 > 0.05 and m3 < 0.5 and \
m4 > 100 and m4 < 200:
return m1 + (m2 * 1. / (1 + e ** (-m3 * (x - m4))))
return (abs(m1) + abs(m2) + abs(m3) + abs(m4)) * 1e14
print curve_fit(f, xdata, ydata)