Hi I have a data set like below df.. I am providing the image and sample dataframe separately.
I want to transform the original dataframe (df) to transformed dataframe (dft) so that I can see the utilisation of each equipment for a 24 hour period (or even longer periods upto 9 days)... at 5 minute intervals. The dft can then be used for plotting... tooltip of the description, etc.
surely, also if you have any alternative simpler solution as opposed to my outline below can also be great too.
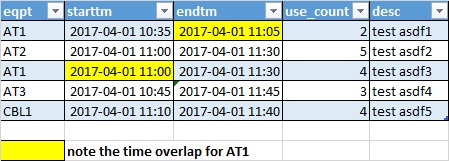
Original Dataframe (df)
Here is the above dataframe (df) which you can copy paste to jupyter to create it:
from io import StringIO
import pandas as pd
dfstr = StringIO(u"""
eqpt;starttm;endtm;use_count;desc
AT1;2017-04-01 10:35;2017-04-01 11:05;2;test asdf1
AT2;2017-04-01 11:00;2017-04-01 11:30;5;test asdf2
AT1;2017-04-01 11:00;2017-04-01 11:30;4;test asdf3
AT3;2017-04-01 10:45;2017-04-01 11:45;3;test asdf4
CBL1;2017-04-01 11:10;2017-04-1 11:40;4;test asdf5
""")
df = pd.read_csv(dfstr, sep=";")
df
I want to transform df to individual rows for each eqpt... with say starttime and endtime from 2017-04-01 00:00 to 23:55 so that I can know the equipment utilisation in each 5 minute grid as well as for plotting and resampling to say maximum in each 1 hour for summary, etc.
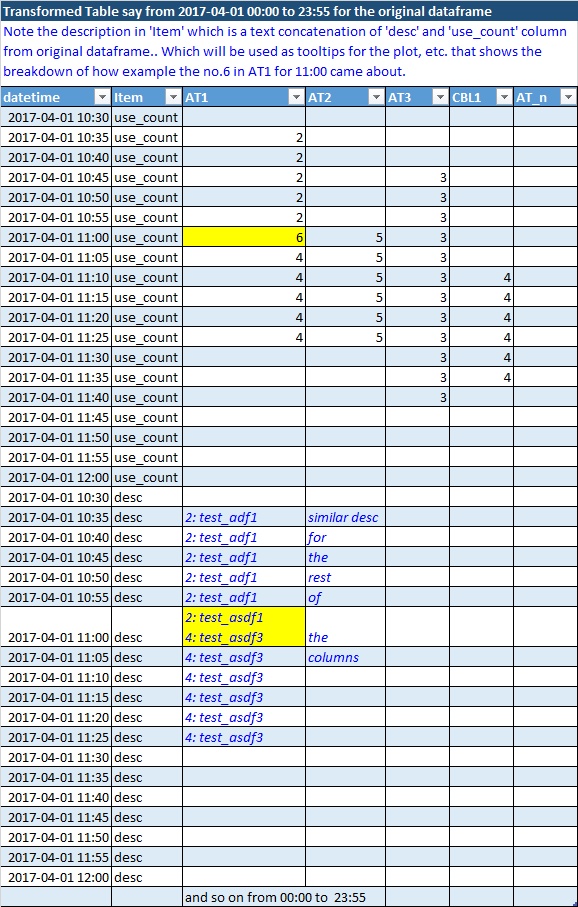
Transformed Dataframe (dft)
Here is the resulting transformed image.. and a sample result dataframe (dft) follows:
columns for this dataframe comes from 'eqpt' of the original dataframe.
just realized that the description column cannot be in the same dataframe dft if need to keep the use_counts aggregate a number only. Thus, please provide any alternate solution that can achieve the same purpose but keeping the columns as float for counts only and description text aggregate elsewhere.. and later can be merged or looked up.
Here is the above dataframe (dft):
dftstr = StringIO(u"""
datetime;Item;AT1;AT2;AT3;CBL1;AT_n
2017-04-01 10:30;use_count;;;;;
2017-04-01 10:35;use_count;2;;;;
2017-04-01 10:40;use_count;2;;;;
2017-04-01 10:45;use_count;2;;3;;
2017-04-01 10:50;use_count;2;;3;;
2017-04-01 10:55;use_count;2;;3;;
2017-04-01 11:00;use_count;6;5;3;;
2017-04-01 11:05;use_count;4;5;3;;
2017-04-01 11:10;use_count;4;5;3;4;
2017-04-01 11:15;use_count;4;5;3;4;
2017-04-01 11:20;use_count;4;5;3;4;
2017-04-01 11:25;use_count;4;5;3;4;
2017-04-01 11:30;use_count;;;3;4;
2017-04-01 11:35;use_count;;;3;4;
2017-04-01 11:40;use_count;;;3;;
2017-04-01 11:45;use_count;;;;;
2017-04-01 11:50;use_count;;;;;
2017-04-01 11:55;use_count;;;;;
2017-04-01 12:00;use_count;;;;;
2017-04-01 10:30;desc;;;;;
2017-04-01 10:35;desc;2: test_adf1;similar desc;;;
2017-04-01 10:40;desc;2: test_adf1;for;;;
2017-04-01 10:45;desc;2: test_adf1;the;;;
2017-04-01 10:50;desc;2: test_adf1;rest;;;
2017-04-01 10:55;desc;2: test_adf1;of;;;
2017-04-01 11:00;desc;"2: test_asdf1
4: test_asdf3";the;;;
2017-04-01 11:05;desc;4: test_asdf3;columns;;;
2017-04-01 11:10;desc;4: test_asdf3;;;;
2017-04-01 11:15;desc;4: test_asdf3;;;;
2017-04-01 11:20;desc;4: test_asdf3;;;;
2017-04-01 11:25;desc;4: test_asdf3;;;;
2017-04-01 11:30;desc;;;;;
2017-04-01 11:35;desc;;;;;
2017-04-01 11:40;desc;;;;;
2017-04-01 11:45;desc;;;;;
2017-04-01 11:50;desc;;;;;
2017-04-01 11:55;desc;;;;;
2017-04-01 12:00;desc;;;;;
;;and so on from 00:00 to 23:55;;;;
""")
dft = pd.read_csv(dftstr, sep=";")
dft