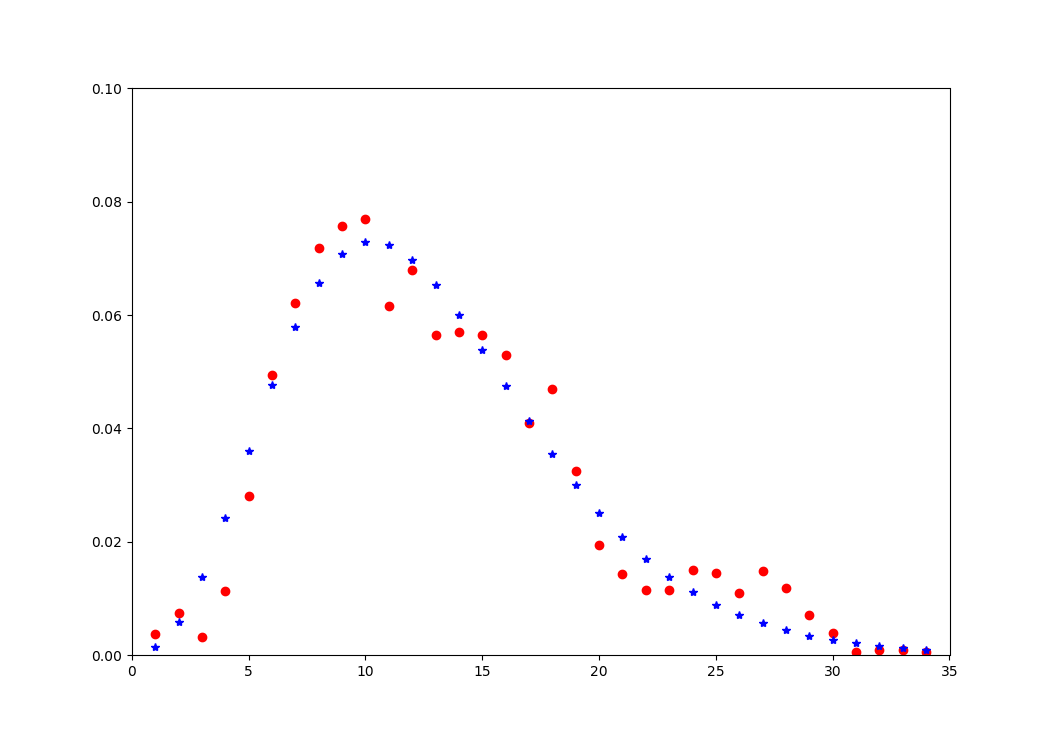
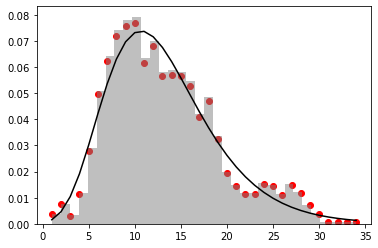
I have a hypothetical y function of x and trying to find/fit a lognormal distribution curve that would shape over the data best. I am using curve_fit function and was able to fit normal distribution, but the curve does not look optimized.
Below are the give y and x data points where y = f(x).
y_axis = [0.00032425299473065838, 0.00063714106162861229, 0.00027009331177605913, 0.00096672396877715144, 0.002388766809835889, 0.0042233337680543182, 0.0053072824980722137, 0.0061291327849408699, 0.0064555344006149871, 0.0065601228278316746, 0.0052574034010282218, 0.0057924488798939255, 0.0048154093097913355, 0.0048619350036057446, 0.0048154093097913355, 0.0045114840997070331, 0.0034906838696562147, 0.0040069911024866456, 0.0027766995669134334, 0.0016595801819374015, 0.0012182145074882836, 0.00098231827111984341, 0.00098231827111984363, 0.0012863691645616997, 0.0012395921040321833, 0.00093554121059032721, 0.0012629806342969417, 0.0010057068013846018, 0.0006081017868837127, 0.00032743942370661445, 4.6777060529516312e-05, 7.0165590794274467e-05, 7.0165590794274467e-05, 4.6777060529516745e-05]
y-axis are probabilities of an event occurring in x-axis time bins:
x_axis = [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 12.0, 13.0, 14.0, 15.0, 16.0, 17.0, 18.0, 19.0, 20.0, 21.0, 22.0, 23.0, 24.0, 25.0, 26.0, 27.0, 28.0, 29.0, 30.0, 31.0, 32.0, 33.0, 34.0]
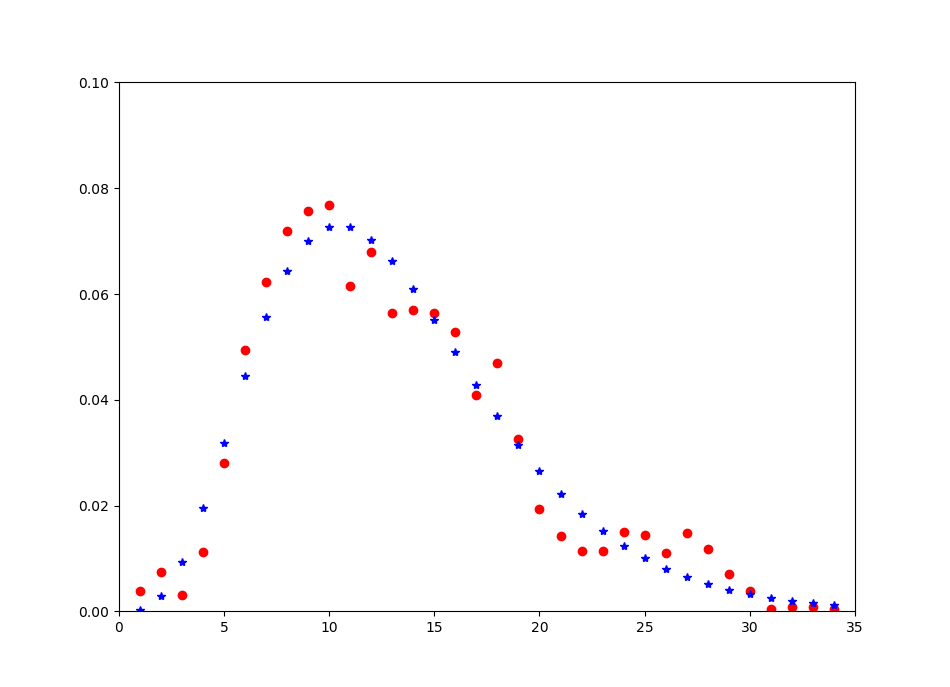
I was able to get a better fit on my data using excel and lognormal approach. When I attempt to use lognormal in python, the fit does not work and I am doing something wrong.
Below is the code I have for fitting a normal distribution, which seems to be the only one that I can fit in python (hard to believe):
#fitting distributino on top of savitzky-golay
%matplotlib inline
import matplotlib
import matplotlib.pyplot as plt
import pandas as pd
import scipy
import scipy.stats
import numpy as np
from scipy.stats import gamma, lognorm, halflogistic, foldcauchy
from scipy.optimize import curve_fit
matplotlib.rcParams['figure.figsize'] = (16.0, 12.0)
matplotlib.style.use('ggplot')
# results from savgol
x_axis = [1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 12.0, 13.0, 14.0, 15.0, 16.0, 17.0, 18.0, 19.0, 20.0, 21.0, 22.0, 23.0, 24.0, 25.0, 26.0, 27.0, 28.0, 29.0, 30.0, 31.0, 32.0, 33.0, 34.0]
y_axis = [0.00032425299473065838, 0.00063714106162861229, 0.00027009331177605913, 0.00096672396877715144, 0.002388766809835889, 0.0042233337680543182, 0.0053072824980722137, 0.0061291327849408699, 0.0064555344006149871, 0.0065601228278316746, 0.0052574034010282218, 0.0057924488798939255, 0.0048154093097913355, 0.0048619350036057446, 0.0048154093097913355, 0.0045114840997070331, 0.0034906838696562147, 0.0040069911024866456, 0.0027766995669134334, 0.0016595801819374015, 0.0012182145074882836, 0.00098231827111984341, 0.00098231827111984363, 0.0012863691645616997, 0.0012395921040321833, 0.00093554121059032721, 0.0012629806342969417, 0.0010057068013846018, 0.0006081017868837127, 0.00032743942370661445, 4.6777060529516312e-05, 7.0165590794274467e-05, 7.0165590794274467e-05, 4.6777060529516745e-05]
## y_axis values must be normalised
sum_ys = sum(y_axis)
# normalize to 1
y_axis = [_/sum_ys for _ in y_axis]
# def gamma_f(x, a, loc, scale):
# return gamma.pdf(x, a, loc, scale)
def norm_f(x, loc, scale):
# print 'loc: ', loc, 'scale: ', scale, "\n"
return norm.pdf(x, loc, scale)
fitting = norm_f
# param_bounds = ([-np.inf,0,-np.inf],[np.inf,2,np.inf])
result = curve_fit(fitting, x_axis, y_axis)
result_mod = result
# mod scale
# results_adj = [result_mod[0][0]*.75, result_mod[0][1]*.85]
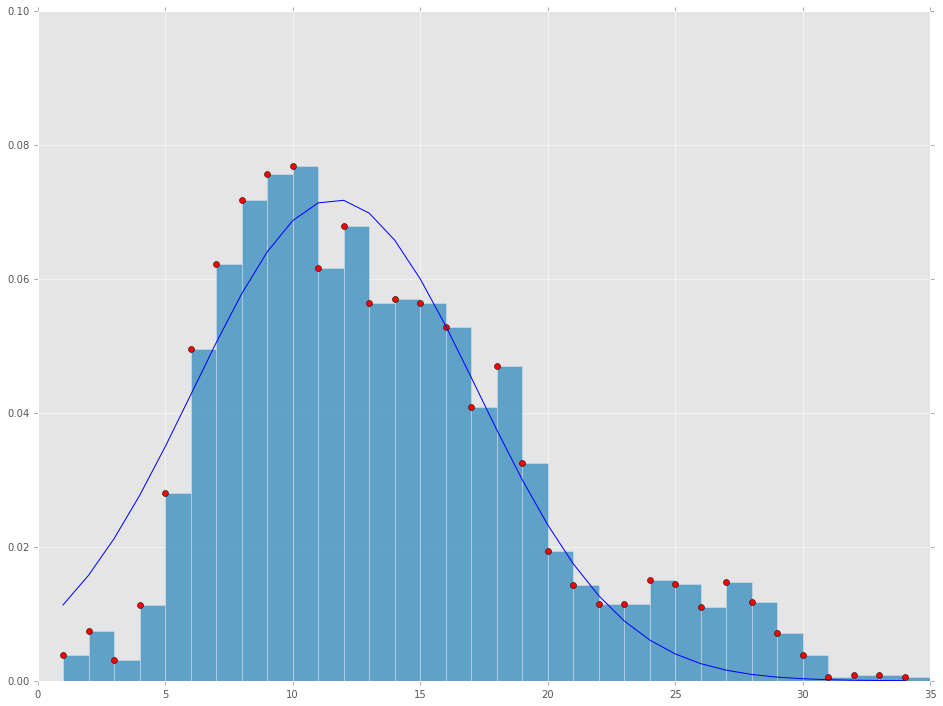
plt.plot(x_axis, y_axis, 'ro')
plt.bar(x_axis, y_axis, 1, alpha=0.75)
plt.plot(x_axis, [fitting(_, *result[0]) for _ in x_axis], 'b-')
plt.axis([0,35,0,.1])
# convert back into probability
y_norm_fit = [fitting(_, *result[0]) for _ in x_axis]
y_fit = [_*sum_ys for _ in y_norm_fit]
print list(y_fit)
plt.show()
I am trying to get answers two questions:
- Is this the best fit I will get from normal distribution curve? How can I imporve my the fit?
- How can I fit a lognormal distribution to this data or is there a better distribution that I can use?
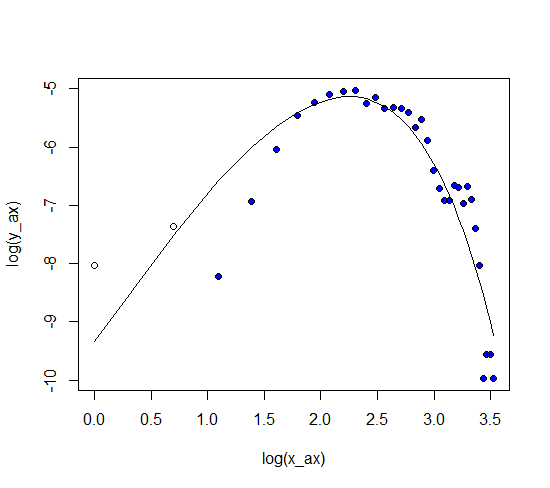
I was playing around with lognormal distribution curve adjust mu and sigma, it looks like that there is possible a better fit. I don't understand what I am doing wrong to get similar results in python.