I frequently find myself calculating summary statistics in R using dplyr, then writing the result to csv and loading it into Tableau to generate a table because Tableau's tables are so simple and easy. I'd much rather generate the tables directly in R.
Is there an easy solution for grouped tables in R?
It's very easy to generate the data I would want:
library(tidyr)
library(dplyr)
summary_table <- iris %>%
gather(measure, value, -Species) %>%
separate(measure, into=c("attribute", "dimension")) %>%
group_by(Species, attribute, dimension) %>%
summarise(mean=mean(value))
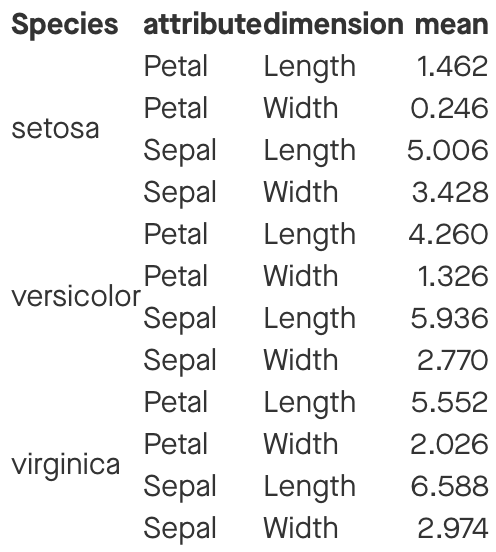
summary_table
Source: local data frame [12 x 4]
Groups: Species, attribute [?]
Species attribute dimension mean
<fctr> <chr> <chr> <dbl>
1 setosa Petal Length 1.462
2 setosa Petal Width 0.246
3 setosa Sepal Length 5.006
4 setosa Sepal Width 3.428
5 versicolor Petal Length 4.260
6 versicolor Petal Width 1.326
7 versicolor Sepal Length 5.936
8 versicolor Sepal Width 2.770
9 virginica Petal Length 5.552
10 virginica Petal Width 2.026
11 virginica Sepal Length 6.588
12 virginica Sepal Width 2.974
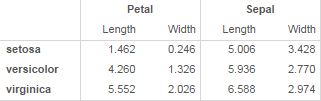
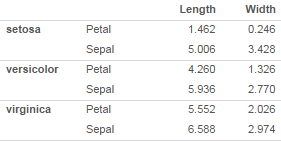
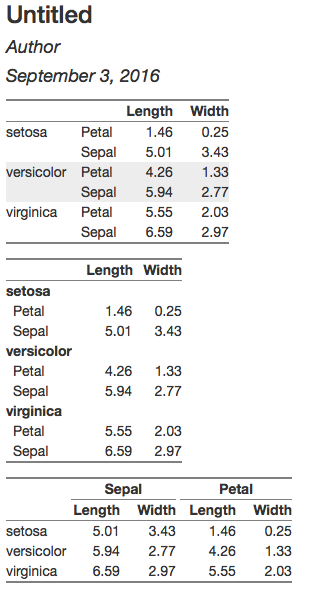
Now I'd like to present this as:
I'd want to try a few different ways of organizing, so I'd want to be able to easily group on rows instead of columns
The key features of the grouped rows version are:
- Grouping variable is on the left, in a separate column rather than a separate row, in a cell that spans all of the rows
- Horizontal cell borders at the group level
I'm new to rmarkdown, but the ultimate goal is to have this in an html doc.
Is this possible?

aggregate(x = iris[, colnames(iris)[ colnames(iris) != "Species" ] ], by = list(iris$Species), FUN = function(y){ ifelse(is.numeric(y),mean(y),NA) } )for a start. – Stunk