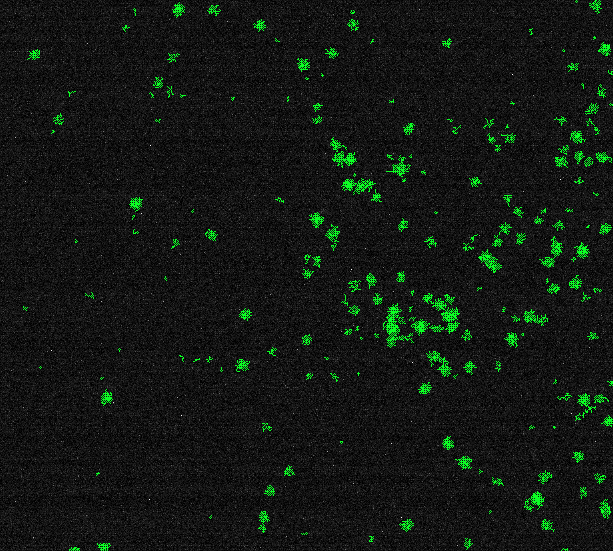
I'm looking for a proper solution how to count particles and measure their sizes in this image:

In the end I have to obtain the lists of particles' coordinates and area squares. After some search on the internet I realized there are 3 approaches for particles detection:
- blobs
- Contours
- connectedComponentsWithStats
Looking at different projects I assembled some code with the mix of it.
import pylab
import cv2
import numpy as np
Gaussian blurring and thresholding
original_image = cv2.imread(img_path)
img = original_image
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
img = cv2.GaussianBlur(img, (5, 5), 0)
img = cv2.blur(img, (5, 5))
img = cv2.medianBlur(img, 5)
img = cv2.bilateralFilter(img, 6, 50, 50)
max_value = 255
adaptive_method = cv2.ADAPTIVE_THRESH_GAUSSIAN_C
threshold_type = cv2.THRESH_BINARY
block_size = 11
img_thresholded = cv2.adaptiveThreshold(img, max_value, adaptive_method, threshold_type, block_size, -3)
filter small objects
min_size = 4
nb_components, output, stats, centroids = cv2.connectedComponentsWithStats(img, connectivity=8)
sizes = stats[1:, -1]
nb_components = nb_components - 1
# for every component in the image, you keep it only if it's above min_size
for i in range(0, nb_components):
if sizes[i] < min_size:
img[output == i + 1] = 0
generation of Contours for filling holes and measurements. pos_list and size_list is what we were looking for
contours, hierarchy = cv2.findContours(img, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
pos_list = []
size_list = []
for i in range(len(contours)):
area = cv2.contourArea(contours[i])
size_list.append(area)
(x, y), radius = cv2.minEnclosingCircle(contours[i])
pos_list.append((int(x), int(y)))
for the self-check, if we plot these coordinates over the original image
pts = np.array(pos_list)
pylab.figure(0)
pylab.imshow(original_image)
pylab.scatter(pts[:, 0], pts[:, 1], marker="x", color="green", s=5, linewidths=1)
pylab.show()
We might get something like the following:

And... I'm not really satisfied with the results. Some clearly visible particles are not included, on the other side, some doubt fluctuations of intensity have been counted. I'm playing now with different filters' settings, but the feeling is it's wrong.
If someone knows how to improve my solution, please share.