dplyr functions work on data.tables, so here's a dplyr solution that also "avoids the for-loop" :)
dt %>% mutate(across(all_of(cols), ~ -1 * .))
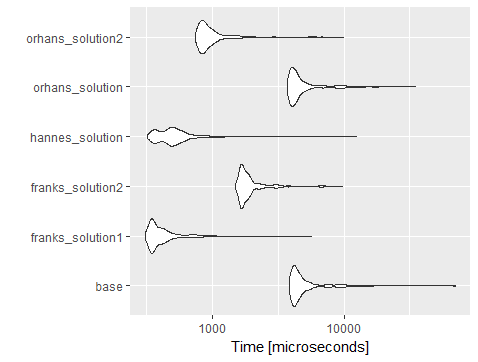
I benchmarked it using orhan's code (adding rows and columns) and you'll see dplyr::mutate with across mostly executes faster than most of the other solutions and slower than the data.table solution using lapply.
library(data.table); library(dplyr)
dt <- data.table(a = 1:100000, b = 1:100000, d = 1:100000) %>%
mutate(a2 = a, a3 = a, a4 = a, a5 = a, a6 = a)
cols <- c("a", "b", "a2", "a3", "a4", "a5", "a6")
dt %>% mutate(across(all_of(cols), ~ -1 * .))
#> a b d a2 a3 a4 a5 a6
#> 1: -1 -1 1 -1 -1 -1 -1 -1
#> 2: -2 -2 2 -2 -2 -2 -2 -2
#> 3: -3 -3 3 -3 -3 -3 -3 -3
#> 4: -4 -4 4 -4 -4 -4 -4 -4
#> 5: -5 -5 5 -5 -5 -5 -5 -5
#> ---
#> 99996: -99996 -99996 99996 -99996 -99996 -99996 -99996 -99996
#> 99997: -99997 -99997 99997 -99997 -99997 -99997 -99997 -99997
#> 99998: -99998 -99998 99998 -99998 -99998 -99998 -99998 -99998
#> 99999: -99999 -99999 99999 -99999 -99999 -99999 -99999 -99999
#> 100000: -100000 -100000 100000 -100000 -100000 -100000 -100000 -100000
library(microbenchmark)
mbm = microbenchmark(
base_with_forloop = for (col in 1:length(cols)) {
dt[ , eval(parse(text = paste0(cols[col], ":=-1*", cols[col])))]
},
franks_soln1_w_lapply = dt[ , (cols) := lapply(.SD, "*", -1), .SDcols = cols],
franks_soln2_w_forloop = for (j in cols) set(dt, j = j, value = -dt[[j]]),
orhans_soln_w_forloop = for (j in cols) dt[,(j):= -1 * dt[, ..j]],
orhans_soln2 = dt[,(cols):= - dt[,..cols]],
dplyr_soln = (dt %>% mutate(across(all_of(cols), ~ -1 * .))),
times=1000
)
library(ggplot2)
ggplot(mbm) +
geom_violin(aes(x = expr, y = time)) +
coord_flip()
![]()
Created on 2020-10-16 by the reprex package (v0.3.0)


setwith afor-loop. I suspect it'll be faster. – Jett