Say I have two options for generating the Adjacency Matrix of a network: nx.adjacency_matrix() and my own code. I wanted to test the correctness of my code and came up with some strange inequalities.
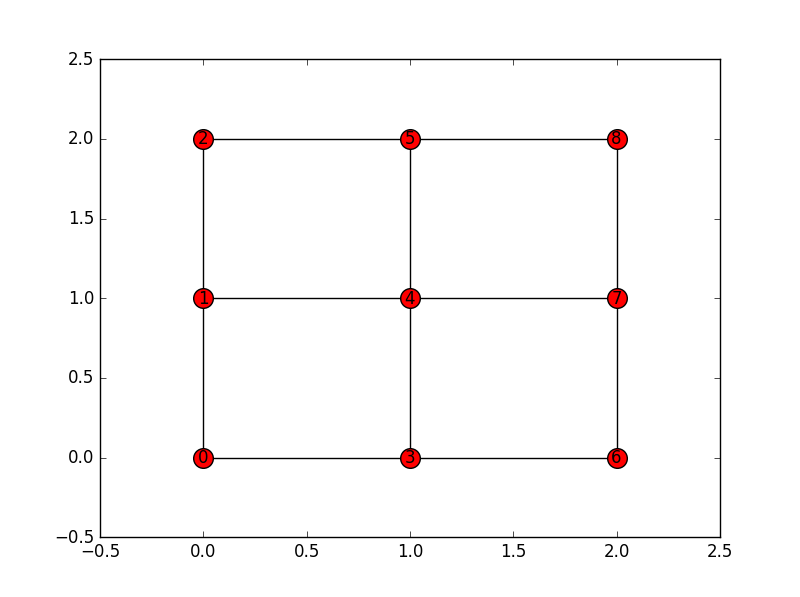
Example: a 3x3 lattice network.
import networkx as nx
N=3
G=nx.grid_2d_graph(N,N)
pos = dict( (n, n) for n in G.nodes() )
labels = dict( ((i,j), i + (N-1-j) * N ) for i, j in G.nodes() )
nx.relabel_nodes(G,labels,False)
inds=labels.keys()
vals=labels.values()
inds.sort()
vals.sort()
pos2=dict(zip(vals,inds))
plt.figure()
nx.draw_networkx(G, pos=pos2, with_labels=True, node_size = 200)
The adjacency matrix with nx.adjacency_matrix():
B=nx.adjacency_matrix(G)
B1=B.todense()
[[0 0 0 0 0 1 0 0 1]
[0 0 0 1 0 1 0 0 0]
[0 0 0 1 0 1 0 1 1]
[0 1 1 0 0 0 1 0 0]
[0 0 0 0 0 0 0 1 1]
[1 1 1 0 0 0 0 0 0]
[0 0 0 1 0 0 0 1 0]
[0 0 1 0 1 0 1 0 0]
[1 0 1 0 1 0 0 0 0]]
According to it, node 0 (entire 1st row and entire 1st column) is connected to nodes 5 and 8. But if you look at the image above this is wrong, as it connects to nodes 1 and 3.
Now my code (to be run in in the same script as the above):
import numpy
import math
P=3
def nodes_connected(i, j):
try:
if i in G.neighbors(j):
return 1
except nx.NetworkXError:
return False
A=numpy.zeros((P*P,P*P))
for i in range(0,P*P,1):
for j in range(0,P*P,1):
if i not in G.nodes():
A[i][:]=0
A[:][i]=0
elif i in G.nodes():
A[i][j]=nodes_connected(i,j)
A[j][i]=A[i][j]
for i in range(0,P*P,1):
for j in range(0,P*P,1):
if math.isnan(A[i][j]):
A[i][j]=0
print(A)
This yields:
[[ 0. 1. 0. 1. 0. 0. 0. 0. 0.]
[ 1. 0. 1. 0. 1. 0. 0. 0. 0.]
[ 0. 1. 0. 0. 0. 1. 0. 0. 0.]
[ 1. 0. 0. 0. 1. 0. 1. 0. 0.]
[ 0. 1. 0. 1. 0. 1. 0. 1. 0.]
[ 0. 0. 1. 0. 1. 0. 0. 0. 1.]
[ 0. 0. 0. 1. 0. 0. 0. 1. 0.]
[ 0. 0. 0. 0. 1. 0. 1. 0. 1.]
[ 0. 0. 0. 0. 0. 1. 0. 1. 0.]]
which says that node 0 is connected to nodes 1 and 3. Why does such difference exist? What is wrong in this situation?

dictabove, you should change to usingcollections.OrderedDict... – Pie