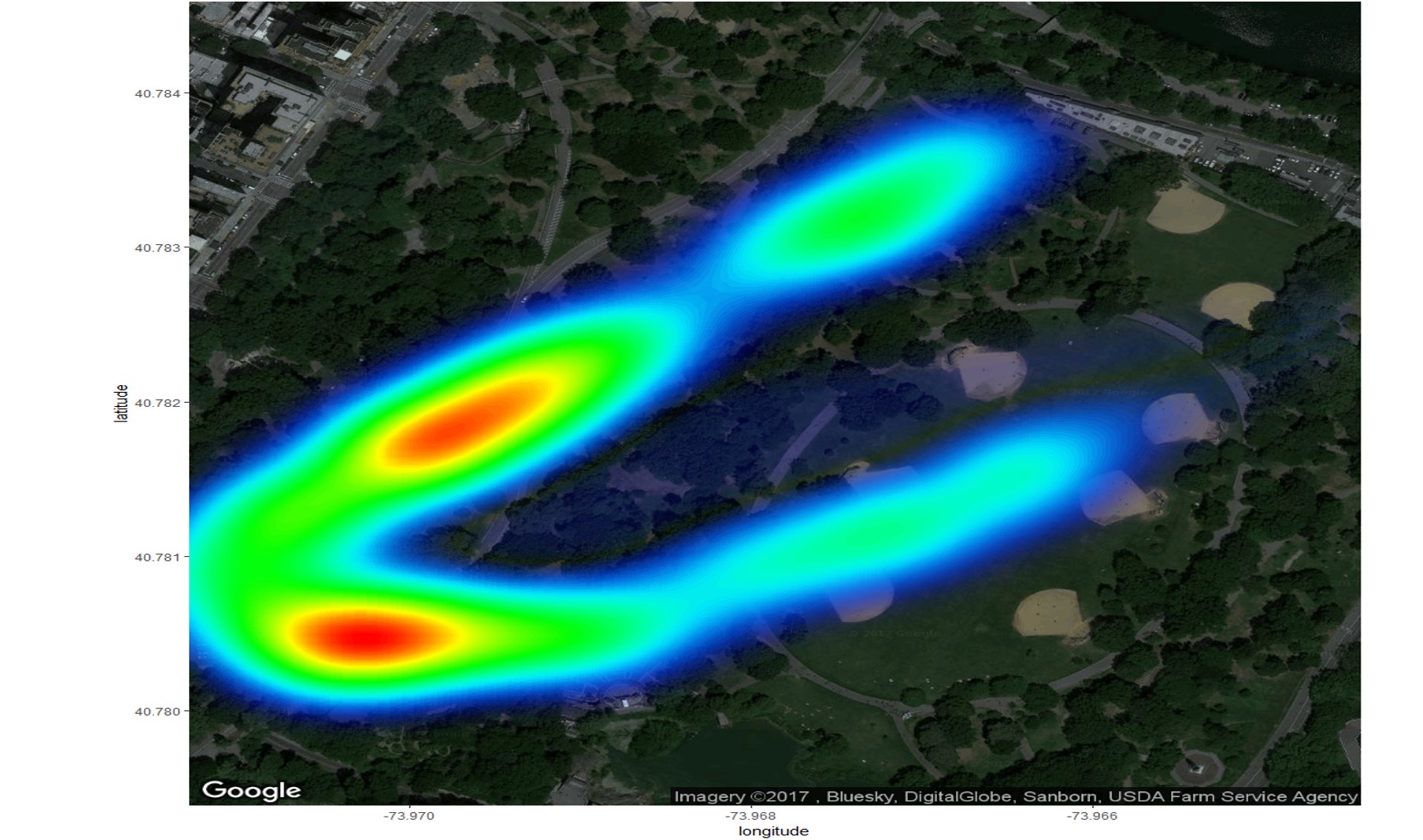
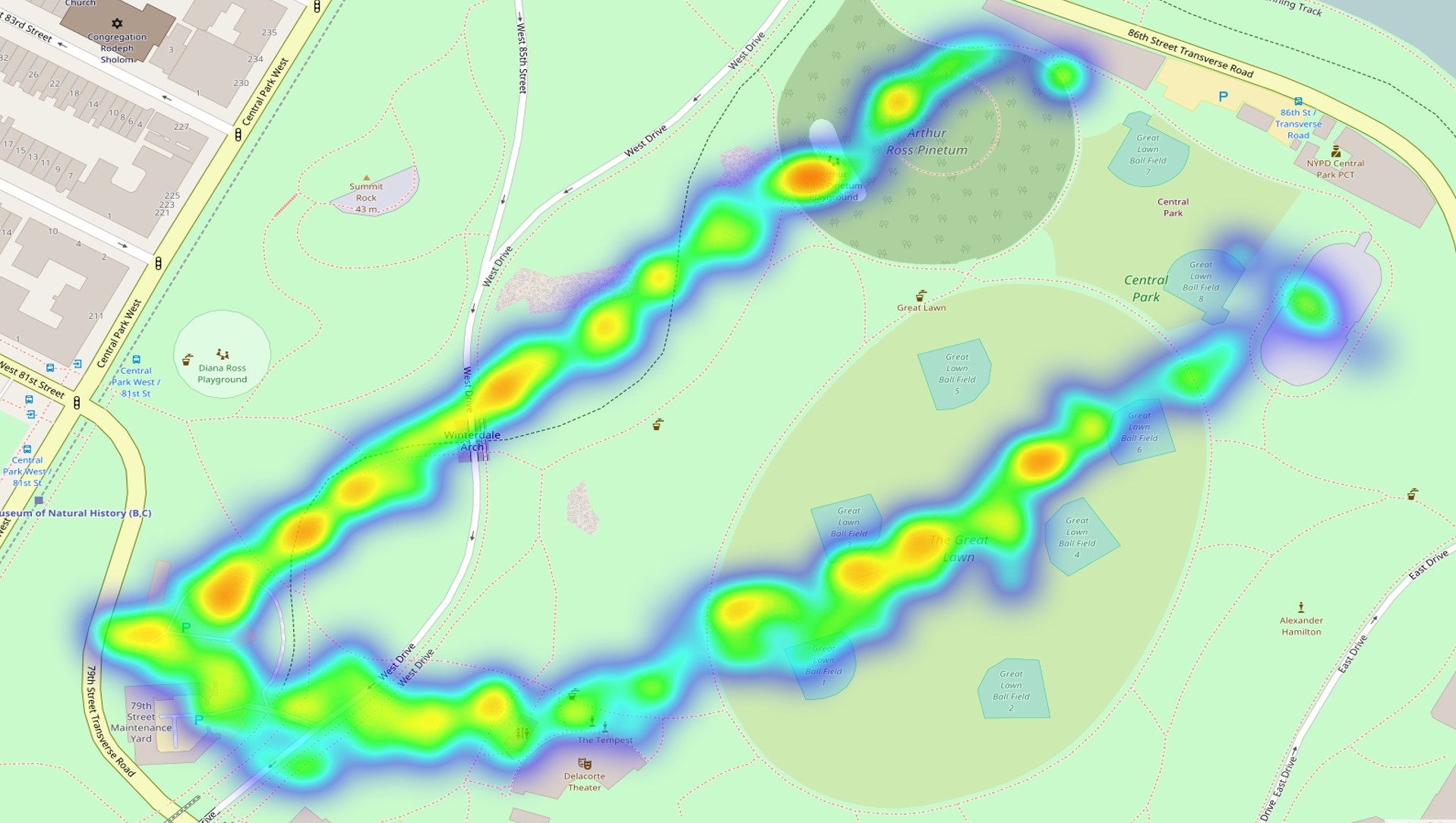
I made static heatmaps with the library(ggmap) and the stat_density2d() function. Looking to recreate this in a shiny app on a dynamic leaflet map, I found addHeatmap(). However, the resulting images are dissimilar, with the ggmap version seemingly offering the correct result.
GGMAP
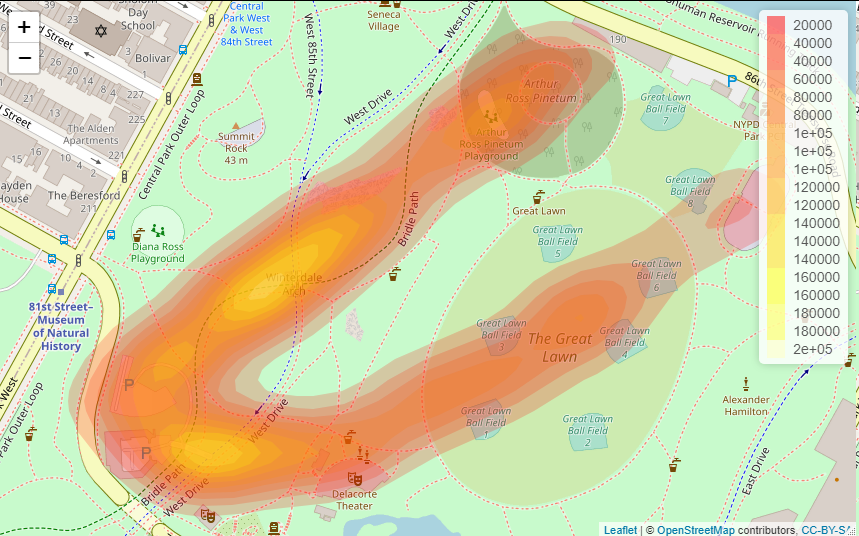
LEAFLET
What is causing this difference?
To run both of the below reproducible examples, you can download some data (csv file) I put here. https://drive.google.com/drive/folders/0B8_GTHBuoKSRR1VIRmhOUTJKYU0?usp=sharing
Note that the leaflet result differs with zoom level, but never matches the ggmap result (e.g. in terms location of maximum heat).
This is the ggmap code.
library(ggmap)
data <- read.csv("DATA.csv", sep=";")
data <- subset(data, !is.na(CrdLatDeg))
xmin <- min(data$CrdLonDeg)
xmax <- max(data$CrdLonDeg)
ymin <- min(data$CrdLatDeg)
ymax <- max(data$CrdLatDeg)
lon <- c(xmin,xmax)
lat <- c(ymin,ymax)
map <- get_map(location = c(lon = mean(lon), lat = mean(lat)), zoom = 17,
maptype = "satellite", source = "google")
ggmap(map) +
labs(x="longitude", y="latitude") +
stat_density2d(data=data, aes(x=CrdLonDeg, y=CrdLatDeg, alpha= ..level.., fill= ..level..), colour=FALSE,
geom="polygon", bins=100) +
scale_fill_gradientn(colours=c(rev(rainbow(100, start=0, end=.7)))) + scale_alpha(range=c(0,.8)) +
guides(alpha=FALSE,fill=FALSE)
This is the leaflet code.
library(leaflet.extras)
data <- read.csv("DATA.csv", sep=";")
data <- subset(data, !is.na(CrdLatDeg))
leaflet(data) %>%
addTiles(group="OSM") %>%
addHeatmap(group="heat", lng=~CrdLonDeg, lat=~CrdLatDeg, max=.6, blur = 60)

stat_density2d()(which isMASS::bandwidth.nrd()), although it does use polygons. – Selaginella