I am trying to use mle() function in MATLAB to estimate the parameters of a 6-parameter custom distribution.
The PDF of the custom distribution is
and the CDF is
where Γ(x,y) and Γ(x) are the upper incomplete gamma function and the gamma function, respectively. α, θ, β, a, b, and c are the parameters of the custom distribution. K is given by
Given a data vector 'data', I want to estimate the parameters α, θ, β, a, b, and c.
So, far I have come up with this code:
data = rand(20000,1); % Since I cannot upload the acutal data, we may use this
t = 0:0.0001:0.5;
fun = @(w,a,b,c) w^(a-1)*(1-w)^(b-1)*exp^(-c*w);
% to estimate the parameters
custpdf = @(data,myalpha,mybeta,mytheta,a,b,c)...
((integral(@(t)fun(t,a,b,c),0,1)^-1)*...
mybeta*...
igamma(myalpha,((mytheta/t)^mybeta)^(a-1))*...
(mytheta/t)^(myalpha*mybeta+1)*...
exp(-(mytheta/t)^mybeta-(c*(igamma(myalpha,(mytheta/t)^mybeta)/gamma(myalpha)))))...
/...
(mytheta*...
gamma(myalpha)^(a+b-1)*...
(gamma(myalpha)-igamma(myalpha,(mytheta/t)^mybeta))^(1-b));
custcdf = @(data,myalpha,mybeta,mytheta,a,b,c)...
(integral(@(t)fun(t,a,b,c),0,1)^-1)*...
integral(@(t)fun(t,a,b,c),0,igamma(myalpha,(mytheta/t)^mybeta)^mybeta/gamma(myalpha));
phat = mle(data,'pdf',custpdf,'cdf',custcdf,'start',0.0);
But I get the following error:
Error using mlecustom (line 166)
Error evaluating the user-supplied pdf function
'@(data,myalpha,mybeta,mytheta,a,b,c)((integral(@(t)fun(t,a,b,c),0,1)^-1)*mybeta*igamma(myalpha,((mytheta/t)^mybeta)^(a-1))*(mytheta/t)^(myalpha*mybeta+1)*exp(-(mytheta/t)^mybeta-(c*(igamma(myalpha,(mytheta/t)^mybeta)/gamma(myalpha)))))/(mytheta*gamma(myalpha)^(a+b-1)*(gamma(myalpha)-igamma(myalpha,(mytheta/t)^mybeta))^(1-b))'.
Error in mle (line 245)
phat = mlecustom(data,varargin{:});
Caused by:
Not enough input arguments.
I tried to look into the error lines but I can't figure out where the error actually is.
Which function lacks fewer inputs? Is it referring to fun? Why would mle lack fewer inputs when it is trying to estimate the parameters?
Could someone kindly help me debug the error?
Thanks in advance.

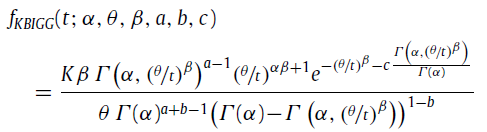
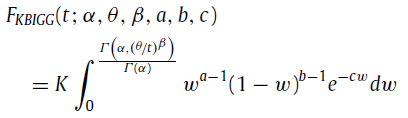
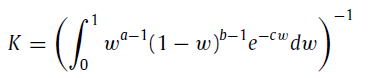
custpdfas thepdfformle, becausemleonly provides 2 inputs (see docs) - "This custom function accepts the vectordataand one or more individual distribution parameters as input parameters, and returns a vector of cumulative probability values.". If you want to pass more than 2 variables to your function, you should do something like what's shown here. Same goes forcustcdf. – Icemanmle(data,'pdf',custpdf,'cdf',custcdf,'start',0.0);should bemle(data,'pdf',@data custpdf(data,myalpha,mybeta,mytheta,a,b,c),'cdf',@data custcdf(data,myalpha,mybeta,mytheta,a,b,c),'start',0.0);– Dipterocarpaceousfunction out = custpdf(...)). Debugging multi-line anonymous functions isn't exactly convenient. – Iceman