First you need to identify the order of the images stores in the 4th dimensions.
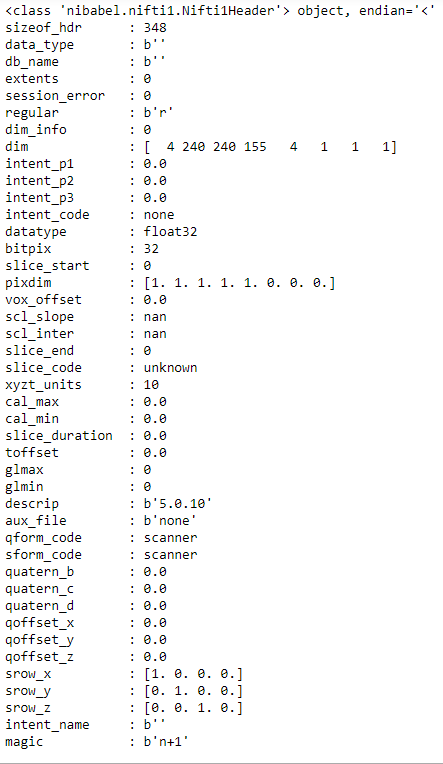
Probably the header will help:
print(a.header)
Next, to keep only 1 modality you can use this:
data = a.get_fdata()
modality_1 = data[:,:,:,0]
EDIT 1:
Based on the website of the challenge:
All BraTS multimodal scans are available as NIfTI files (.nii.gz) and
describe a) native (T1) and b) post-contrast T1-weighted (T1Gd), c)
T2-weighted (T2), and d) T2 Fluid Attenuated Inversion Recovery
(FLAIR) volumes, and were acquired with different clinical protocols
and various scanners from multiple (n=19) institutions, mentioned as
data contributors here.
and
The provided data are distributed after their pre-processing, i.e.
co-registered to the same anatomical template, interpolated to the
same resolution (1 mm^3) and skull-stripped.
So the header will not help in this case (equal dimensions for all modalities due to preprocessing).
If you are looking for the post-contrast T1-weighted (T1Gd) images then it's the 2nd dimension so use:
data = a.get_fdata()
modality_1 = data[:,:,:,1]
Additionally, we can visualize the each 3D volume (data[:,:,:,0], data[:,:,:,1],data[:,:,:,2], data[:,:,:,3]) and verify my statement.
See here: https://gofile.io/?c=fhoZTu

a– Perishablea. tryprint(a.header)– Perishable