My optimization task deals with calculation of the following integral and finding the best values of xl and xu:
Iterations take too long, so I decided to speed them up by calculating integral for all possible values xl and xu and then interpolate calculated values during optimization.
I wrote the following function:
def k_integrand(x, xl, xu):
return((x**2)*mpmath.exp(x))/((xu - xl)*(mpmath.exp(x)-1)**2)
@np.vectorize
def K(xl, xu):
y, err = integrate.quad(k_integrand, xl, xu, args = (xl, xu))
return y
and two identical arrays grid_xl and grid_xu with dinamic increment of values.
When I run the code I get this:
K(grid_xl, grid_xu)
Traceback (most recent call last):
File "<ipython-input-2-5b9df02f12b7>", line 1, in <module>
K(grid_xl, grid_xu)
File "C:/Users/909404/OneDrive/Работа/ZnS-FeS/Теплоемкость/Python/CD357/4 - Optimization CD357 interpolation.py", line 75, in K
y, err = integrate.quad(k_integrand, xl, xu, args = (xl, xu))
File "C:\Users\909404\Anaconda3\lib\site-packages\scipy\integrate\quadpack.py", line 323, in quad
points)
File "C:\Users\909404\Anaconda3\lib\site-packages\scipy\integrate\quadpack.py", line 372, in _quad
if (b != Inf and a != -Inf):
ValueError: The truth value of an array with more than one element is ambiguous. Use a.any() or a.all()
I guess it comes from the fact that xl should be always less than xu.
Is there any way to compare the values of xl and xu and return NaN in case if xl>=xu?
In the end I want to have something like this:
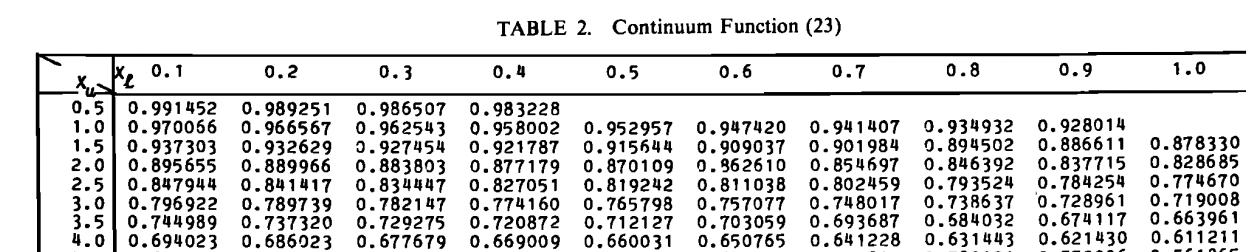
And to have the ability to use interpolation.
Maybe I have chosen the wrong way? I'd appreciate any help.

aandbin your case? Providing the full error traceback would be quite helpful. – Ascending(b != Inf and a != -Inf)is not possible to evaluate withbandabeing arrays of multiple values. Note the source code here, on line372onwards; the code decides to use different integration methods depending on whether the boundsaandbis finite or not. In general, unless the_quadpackroutines are shown to accept vector arguments, your approach won't work. The parameter description on line75and77specifically says thataandbneed to befloats. – Ascending