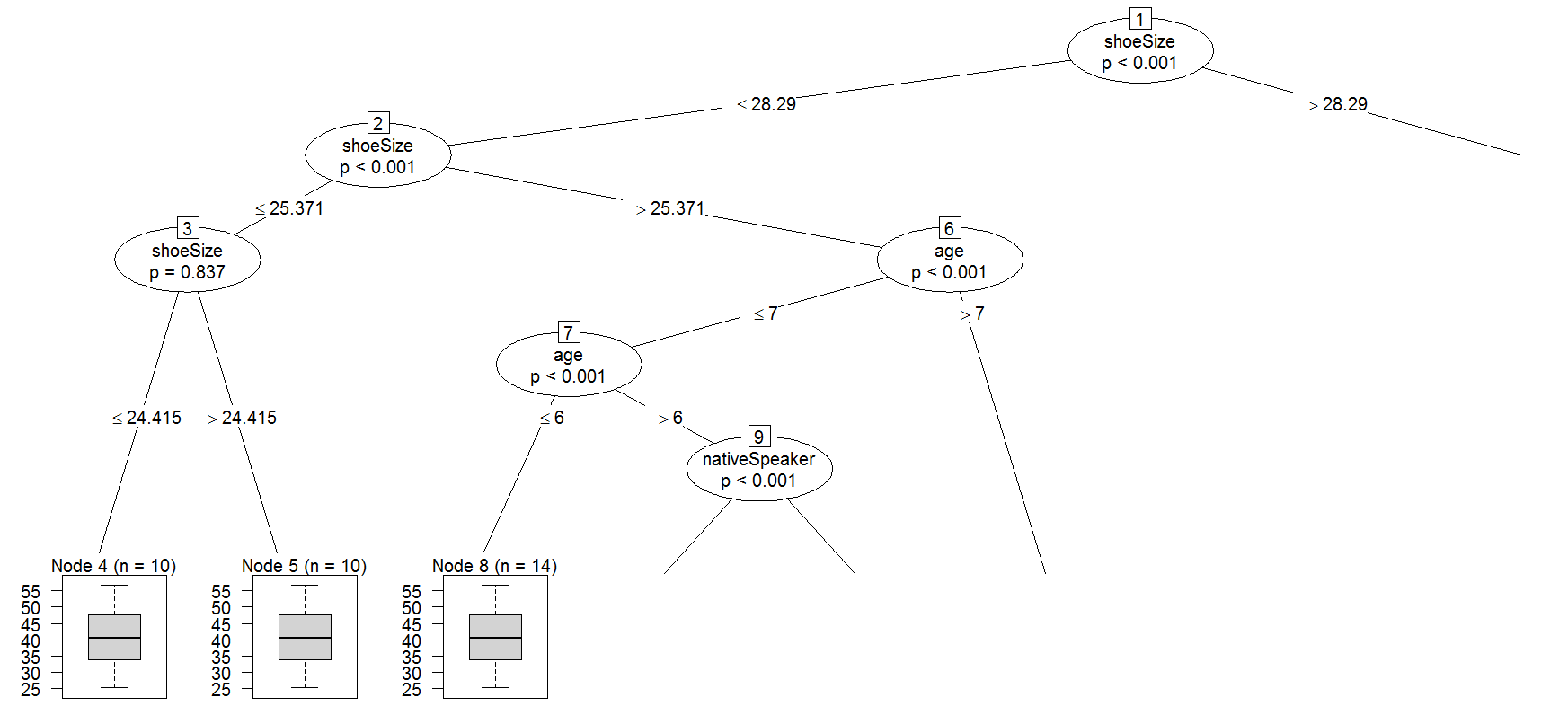
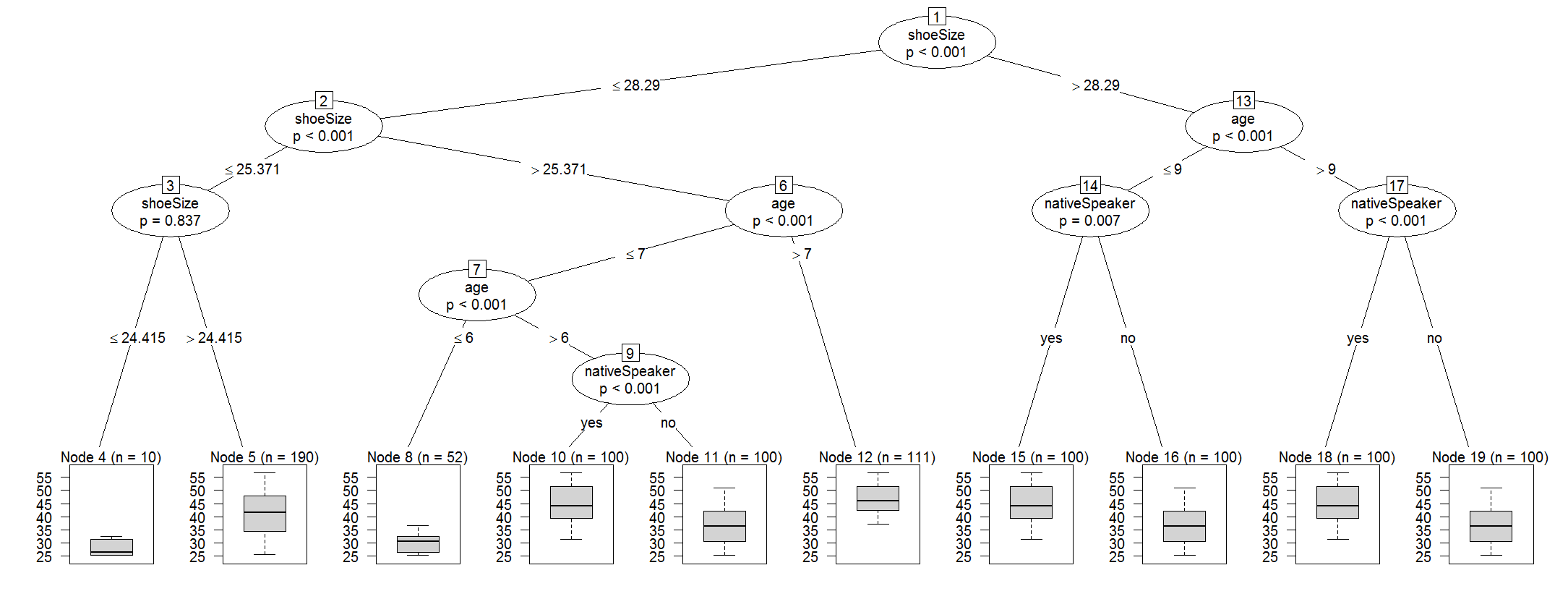
I'm trying to use cforest function(R, party package).
This's what I do to construct forest:
library("party")
set.seed(42)
readingSkills.cf <- cforest(score ~ ., data = readingSkills,
control = cforest_unbiased(mtry = 2, ntree = 50))
Then I want to print the first tree and I do
party:::prettytree(readingSkills.cf@ensemble[[1]],names(readingSkills.cf@data@get("input")))
The result look like this
1) shoeSize <= 28.29018; criterion = 1, statistic = 89.711
2) age <= 6; criterion = 1, statistic = 48.324
3) age <= 5; criterion = 0.997, statistic = 8.917
4)* weights = 0
3) age > 5
5)* weights = 0
2) age > 6
6) age <= 7; criterion = 1, statistic = 13.387
7) shoeSize <= 26.66743; criterion = 0.214, statistic = 0.073
8)* weights = 0
7) shoeSize > 26.66743
9)* weights = 0
6) age > 7
10)* weights = 0
1) shoeSize > 28.29018
11) age <= 9; criterion = 1, statistic = 36.836
12) nativeSpeaker == {}; criterion = 0.998, statistic = 9.347
13)* weights = 0
12) nativeSpeaker == {}
14)* weights = 0
11) age > 9
15) nativeSpeaker == {}; criterion = 1, statistic = 19.124
16) age <= 10; criterion = 1, statistic = 18.441
17)* weights = 0
16) age > 10
18)* weights = 0
15) nativeSpeaker == {}
19)* weights = 0
Why is it empty(weights in each node is equal to zero)?