Before we start and dive deeper into any hunt for nanoseconds ( and right, it will soon start, as each ns matters as the scaling opens the whole Pandora Box of the problems ), let's agree on the scales - most easy and often "cheap" premature tricks may and often will derail your dreams once the scales of the problem size have grown into realistic scales - the thousands (seen above in both iterators) behave way different for in-cache computing with < 0.5 [ns] data-fetches, than once having grown beyond the L1/L2/L3-cache-sizes for scales above 1E+5, 1E+6, 1E+9, above [GB]s, where each mis-aligned fetch is WAY more EXPENSIVE, than a few 100 [ns]
Q : "... because I have 8 cores, I want to use them to get 8 times faster"
I wish you could, indeed. Yet, sorry for telling the truth straight, the world does not work this way.
See this interactive tool, it will show you both the speedup limits and their principal dependence on the actual production costs of the real-world scaling of the initial problem, as it grows from trivial sizes and these combined effects at scale just click-it and play with the sliders to see it live, in action :
![Animated UI-interactive Law of Diminishing Returns, re-formulated so as to also contain processing add-on setup / termination overheads]()
Q : (is)Pool.map() really passing the content of those big sub list around processes which cause additional copy?
Yes,
it must do so, by design
plus it does that by passing all that data "through" another "expensive" SER/DES processing,
so as to make it happen delivered "there".
The very same would apply vice-versa whenever you would have tried to return "back" some mastodon-sized result(s), which you did not, here above.
Q : But if so, why doesn't it passing the address of sub-list?
Because the remote ( parameter-receiving ) process is another, fully autonomous process, with its own, separate and protected, address-space we cannot just pass an address-reference "into", and we wanted that to be a fully independent, autonomously working python process ( due to a will to use this trick so as to escape from GIL-lock dancing ), didn't we? Sure we did - this is a central step of our escape from the GIL-Wars ( for better understanding of the GIL-lock pros and cons, may like this and this ( Pg.15+ on CPU-bound processing ).
0.1 ns - NOP
0.3 ns - XOR, ADD, SUB
0.5 ns - CPU L1 dCACHE reference (1st introduced in late 80-ies )
0.9 ns - JMP SHORT
1 ns - speed-of-light (a photon) travel a 1 ft (30.5cm) distance -- will stay, throughout any foreseeable future :o)
?~~~~~~~~~~~ 1 ns - MUL ( i**2 = MUL i, i )~~~~~~~~~ doing this 1,000 x is 1 [us]; 1,000,000 x is 1 [ms]; 1,000,000,000 x is 1 [s] ~~~~~~~~~~~~~~~~~~~~~~~~~
3~4 ns - CPU L2 CACHE reference (2020/Q1)
5 ns - CPU L1 iCACHE Branch mispredict
7 ns - CPU L2 CACHE reference
10 ns - DIV
19 ns - CPU L3 CACHE reference (2020/Q1 considered slow on 28c Skylake)
71 ns - CPU cross-QPI/NUMA best case on XEON E5-46*
100 ns - MUTEX lock/unlock
100 ns - own DDR MEMORY reference
135 ns - CPU cross-QPI/NUMA best case on XEON E7-*
202 ns - CPU cross-QPI/NUMA worst case on XEON E7-*
325 ns - CPU cross-QPI/NUMA worst case on XEON E5-46*
10,000 ns - Compress 1K bytes with a Zippy PROCESS
20,000 ns - Send 2K bytes over 1 Gbps NETWORK
250,000 ns - Read 1 MB sequentially from MEMORY
500,000 ns - Round trip within a same DataCenter
?~~~ 2,500,000 ns - Read 10 MB sequentially from MEMORY~~(about an empty python process to copy on spawn)~~~~ x ( 1 + nProcesses ) on spawned process instantiation(s), yet an empty python interpreter is indeed not a real-world, production-grade use-case, is it?
10,000,000 ns - DISK seek
10,000,000 ns - Read 1 MB sequentially from NETWORK
?~~ 25,000,000 ns - Read 100 MB sequentially from MEMORY~~(somewhat light python process to copy on spawn)~~~~ x ( 1 + nProcesses ) on spawned process instantiation(s)
30,000,000 ns - Read 1 MB sequentially from a DISK
?~~ 36,000,000 ns - Pickle.dump() SER a 10 MB object for IPC-transfer and remote DES in spawned process~~~~~~~~ x ( 2 ) for a single 10MB parameter-payload SER/DES + add an IPC-transport costs thereof or NETWORK-grade transport costs, if going into [distributed-computing] model Cluster ecosystem
150,000,000 ns - Send a NETWORK packet CA -> Netherlands
| | | |
| | | ns|
| | us|
| ms|
Q : " what is the correct way to keep parallel scaling when parallel mapping some operations on a list of large things? "
![enter image description here]()
A )
UNDERSTAND THE WAYS TO AVOID OR AT LEAST REDUCE EXPENSES :
Understand all the types of the costs you have to pay and will pay :
spend as low process instantiation costs as possible (rather expensive ) best as a one-time cost only
On macOS, the spawn start method is now the default. The fork start method should be considered unsafe as it can lead to crashes of the subprocess. See bpo-33725.
spend as small amount of costs of parameter-passing as you must ( yes, best avoid repetitive passing those "large things" as parameters )
never waste resources on things that do not perform your job - ( never spawn more processes than was reported by len( os.sched_getaffinity( 0 ) ) - any process more than this will but wait for its next CPU-core-slot, and will but evict other, cache-efficient process, thus re-paying all the fetch-costs once already paid to re-fetch again all data so to camp-em back in-cache for a soon to get evicted again in-cache computing, while those processes that worked so far this way were right evicted (for what good?) by a naive use of as many as multiprocessing.cpu_count()-reported processes, so expensively spawned in the initial Pool-creation )
better re-use a pre-allocated memory, than keep spending ad-hoc memory allocation costs ALAP
never share a bit, if The Performance is the goal
never block, never - be it python gc which may block if not avoided, or Pool.map() which blocks either
B )
UNDERSTAND THE WAYS TO INCREASE THE EFFICIENCY :
Understand all efficiency increasing tricks, even at a cost of complexity of code ( a few SLOC-s are easy to show in school-books, yet sacrificing both the efficiency and the performance - in spite of these both being your main enemy in a fight for a sustainable performance throughout the scaling ( of either of problem size or iteration depths, or when growing both of them at the same time ).
Some categories of the real-world costs from A ) have dramatically changed the limits of the theoretically achievable speedups to be expected from going into some form of [PARALLEL] process orchestrations ( here, making some parts of the code-execution got executed in the spawned sub-processes ), the initial view of which was first formulated by Dr. Gene Amdahl as early as 60+ years ago ( for which there were recently added two principal extensions of both the process instantiation(s) related setup + termination add on costs ( extremely important in py2 always & py3.5+ for MacOS and Windows ) and an atomicity-of-work, which will be discussed below.
Overhead-strict re-formulation of the Amdahl's Law speedup S:
S = speedup which can be achieved with N processors
s = a proportion of a calculation, which is [SERIAL]
1-s = a parallelizable portion, that may run [PAR]
N = a number of processors ( CPU-cores ) actively participating on [PAR] processing
1
S = __________________________; where s, ( 1 - s ), N were defined above
( 1 - s ) pSO:= [PAR]-Setup-Overhead add-on cost/latency
s + pSO + _________ + pTO pTO:= [PAR]-Terminate-Overhead add-on cost/latency
N
Overhead-strict and resources-aware re-formulation:
1 where s, ( 1 - s ), N
S = ______________________________________________ ; pSO, pTO
| ( 1 - s ) | were defined above
s + pSO + max| _________ , atomicP | + pTO atomicP:= a unit of work,
| N | further indivisible,
a duration of an
atomic-process-block
Prototype on target CPU/RAM device with your python, scaled >>1E+6
Any simplified mock-up example will somehow skew your expectations about how the actual workloads will perform in-vivo. Underestimated RAM-allocations, not seen at small-scales may later surprise at scale, sometimes even throwing the operating system into sluggish states, swapping and thrashing. Some smarter tools ( numba.jit() ) may even analyze the code and shortcut some passages of code, that will never be visited or that does not produce any result, so be warned that simplified examples may lead to surprising observations.
from multiprocessing import Pool
import numpy as np
import os
SCALE = int( 1E9 )
STEP = int( 1E1 )
aLIST = np.random.random( ( 10**3, 10**4 ) ).tolist()
#######################################################################################
# func() does some SCALE'd amount of work, yet
# passes almost zero bytes as parameters
# allocates nothing, but iterator
# returns one byte,
# invariant to any expensive inputs
def func( x ):
for i in range( SCALE ):
i**2
return 1
A few hints on making the strategy of scaling less overhead-costs expensive :
#####################################################################################
# more_work_en_block() wraps some SCALE'd amount of work, sub-list specified
def more_work_en_block( en_block = [ None, ] ):
return [ func( nth_item ) for nth_item in en_block ]
If indeed must pass a big list, better pass larger block, with remote-iterating its parts ( instead of paying transfer-costs for each and every item passed many many more times, than if using sub_blocks ( parameters get SER/DES processed ( ~ the costs of pickle.dumps() + pickle.loads() ) [per-each-call], again, at an add-on costs, that decrease the resulting efficiency and worsen the overheads part of the extended, overhead-strict Amdahl's Law )
#####################################################################################
# some_work_en_block() wraps some SCALE'd amount of work, tuple-specified
def some_work_en_block( sub_block = ( [ None, ], 0, 1 ) ):
return more_work_en_block( en_block = sub_block[0][sub_block[1]:sub_block[2]] )
Right-sizing the number of process-instances :
aMaxNumOfProcessesThatMakesSenseToSPAWN = len( os.sched_getaffinity( 0 ) ) # never more
with Pool( aMaxNumOfProcessesThatMakesSenseToSPAWN ) as p:
p.imap_unordered( more_work_en_block, [ ( aLIST,
start,
start + STEP
)
for start in range( 0, len( aLIST ), STEP ) ] )
Last but not least, expect immense performance boosts from smart use of numpy smart vectorised code, best without repetitive passing of static, pre-copied (during the process instantiation(s), thus paid as the reasonably scaled, here un-avoidable, cost of thereof ) BLOBs, used in the code without passing the same data via parameter-passing, in a vectorised ( CPU-very-efficient ) fashion as read-only data. Some examples on how one can make ~ +500 x speedup one may read here or here, about but ~ +400 x speedup or about a case of just about a ~ +100 x speedup, with some examples of some problem-isolation testing scenarios.
Anyway, the closer will the mock-up code be to your actual workloads, the more sense the benchmarks will get to have ( at scale & in production ).

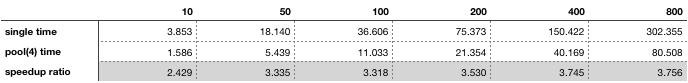
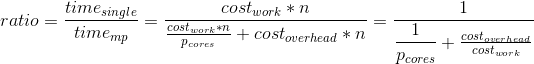
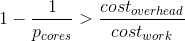
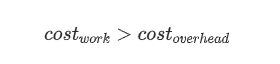

%timeit? – Banburyp.mapworks: Your first example sends a single float to a process 10k times; your second example sends a lists with 10k floats to a process one thousand times; your third example sends a lists with 10k floats to a process ten thousand times. I'm not sure it is fair to compare the second to the first and comparing the third to the second you an 8x increase in time for sending ten times more data - or said another way you get an 8x increase in time for launching ten times more processes. – Banburyfuncreturn just number 1 and add addtional marks. If p.map is really passing large sub list, that will explain the timing. But is there a better parallel approach that can skip passing sub list and directly use data on the main process? I am afraid Thread parallel is not gona work, because python has GIL – Cavefish