This is an interesting question and it would be fun to play approx or approxfun with it
> k <- cumsum(abs(c(x[1], diff(x))))
> c(if (min(k) > 1) seq(min(k) - 1), approxfun(k, x)(min(k):max(k)))
[1] 1 2 3 4 5 4 3 2 1 2 3 4 5 6
Benchmarking (base R options)
(there are some interesting findings, see the result below)
Just borrow the benchmarking example from 989. Given the presented approaches as below
f_Frank <- function(x) Reduce(function(y, z) c(head(y, -1), tail(y, 1):z), x, init = 1L)
f_989_1 <- function(x) c(1, unlist(mapply(function(s, e) tail(s:e, -1), head(c(1, x), -1), x)))
f_989_2 <- function(x) {
c(
seq(x[1] - 1),
unlist(sapply(seq(length(x) - 1), function(i) head(x[i]:x[i + 1], -1))),
tail(x, 1)
)
}
f_akrun <- function(x) {
v1 <- rle(unlist(Map(":", x[-length(x)], x[-1])))$values
c(seq(v1[1]), v1[-1])
}
f_TIC <- function(x) {
k <- cumsum(abs(c(x[1], diff(x))))
c(if (min(k) > 1) seq(min(k) - 1), approxfun(k, x)(min(k):max(k)))
}
we run a vector x of length 500
set.seed(1)
x <- sample(1000, 500, replace = FALSE)
bm <- microbenchmark(
f_Frank(x),
f_989_1(x),
f_989_2(x),
f_akrun(x),
f_TIC(x),
check = "equal"
)
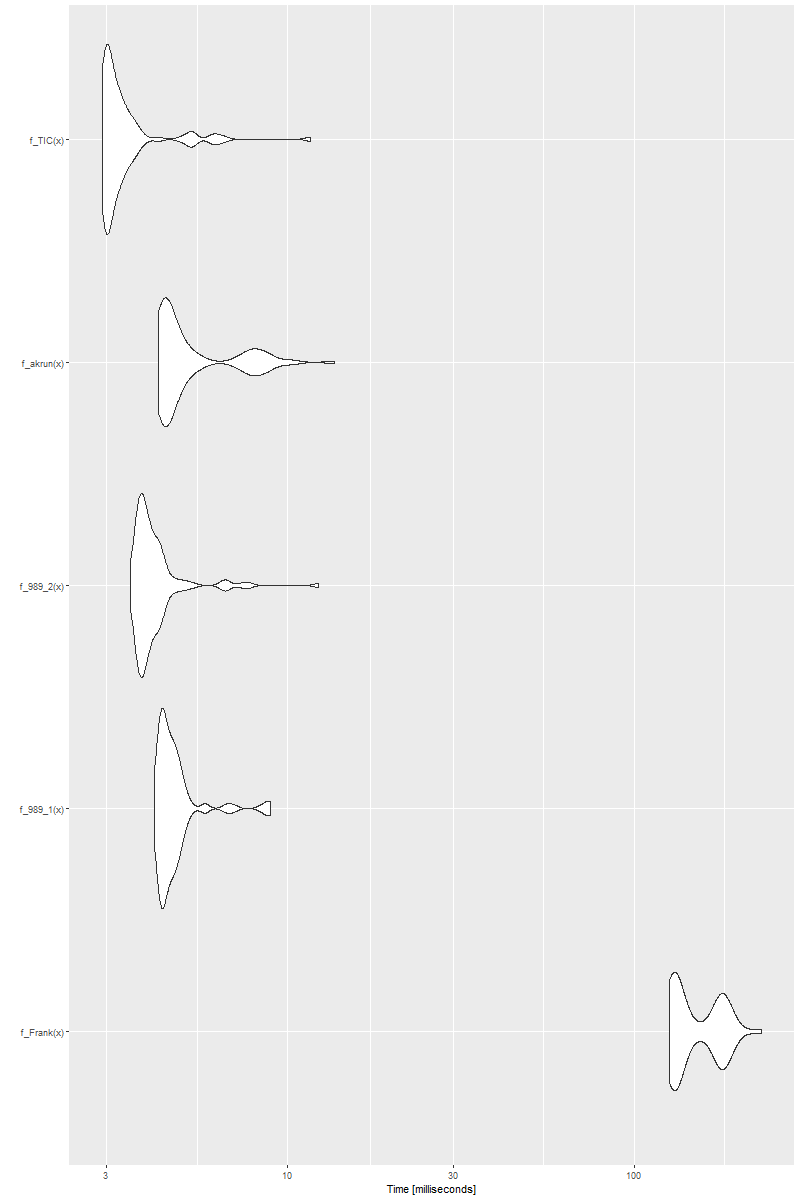
ggplot2::autoplot(bm)
and we will see
![enter image description here]()
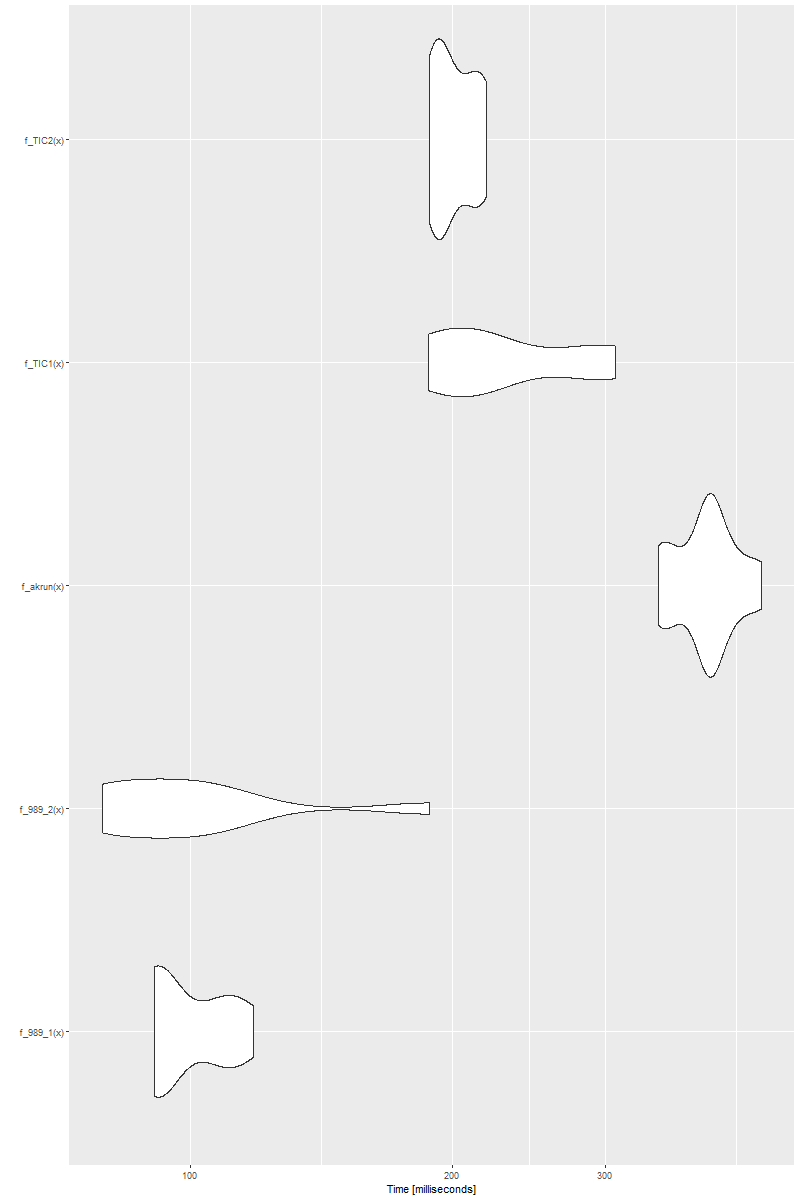
However and interestingly, if we increase the length of x to even longer, say, 5000 for example, i.e., x <- sample(5000, replace = FALSE), we see
![enter image description here]()

1at the beginning, right? – Ironbarksample(x, replace = T)will generate numbers from the minimum and maximum range ofx. Although, is that string the exact outcome you want generated? Or in general something that has a similar functionality tosample? – Conscioussapply...don't know why it was downvoted but:c(1, unlist(sapply(1:(length(x)-1), function(i) head((x[i]:x[i+1]),-1) )), tail(x,1))– Strepphon