I'm trying to plot patient flows between 3 clusters in a Sankey diagram. I have a pd.DataFrame counts with from-to values, see below. To reproduce this DF, here is the counts dict that should be loaded into a pd.DataFrame (which is the input for the visualize_cluster_flow_counts function).
from to value
0 C1_1 C1_2 867
1 C1_1 C2_2 405
2 C1_1 C0_2 2
3 C2_1 C1_2 46
4 C2_1 C2_2 458
... ... ... ...
175 C0_20 C0_21 130
176 C0_20 C2_21 1
177 C2_20 C1_21 12
178 C2_20 C0_21 0
179 C2_20 C2_21 96
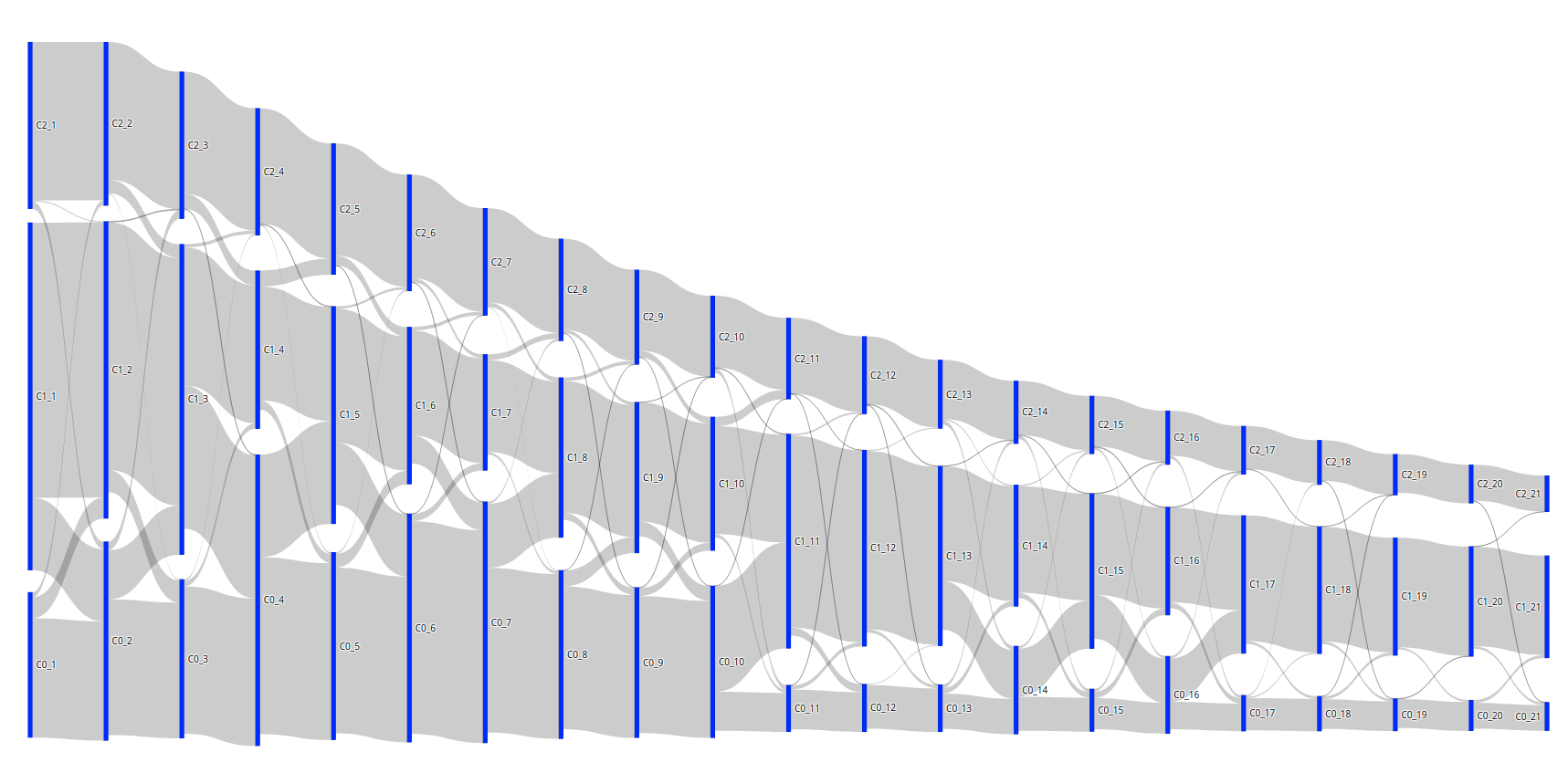
The from and to values in the DataFrame represent the cluster number (either 0, 1, or 2) and the amount of days for the x-axis (between 1 and 21). If I plot the Sankey diagram with these values, this is the result:
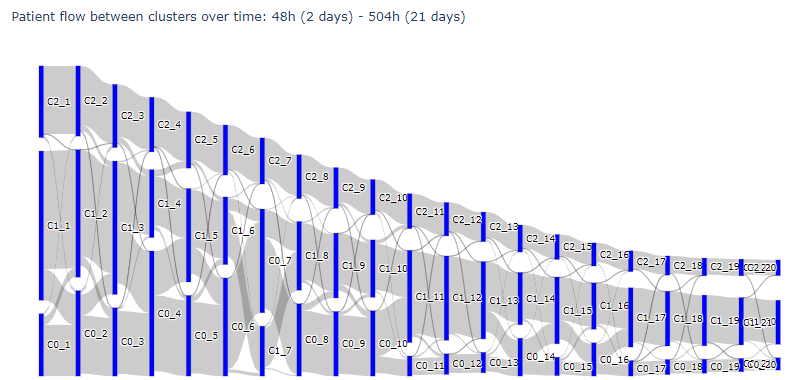
Code:
import plotly.graph_objects as go
def visualize_cluster_flow_counts(counts):
all_sources = list(set(counts['from'].values.tolist() + counts['to'].values.tolist()))
froms, tos, vals, labs = [], [], [], []
for index, row in counts.iterrows():
froms.append(all_sources.index(row.values[0]))
tos.append(all_sources.index(row.values[1]))
vals.append(row[2])
labs.append(row[3])
fig = go.Figure(data=[go.Sankey(
arrangement='snap',
node = dict(
pad = 15,
thickness = 5,
line = dict(color = "black", width = 0.1),
label = all_sources,
color = "blue"
),
link = dict(
source = froms,
target = tos,
value = vals,
label = labs
))])
fig.update_layout(title_text="Patient flow between clusters over time: 48h (2 days) - 504h (21 days)", font_size=10)
fig.show()
visualize_cluster_flow_counts(counts)
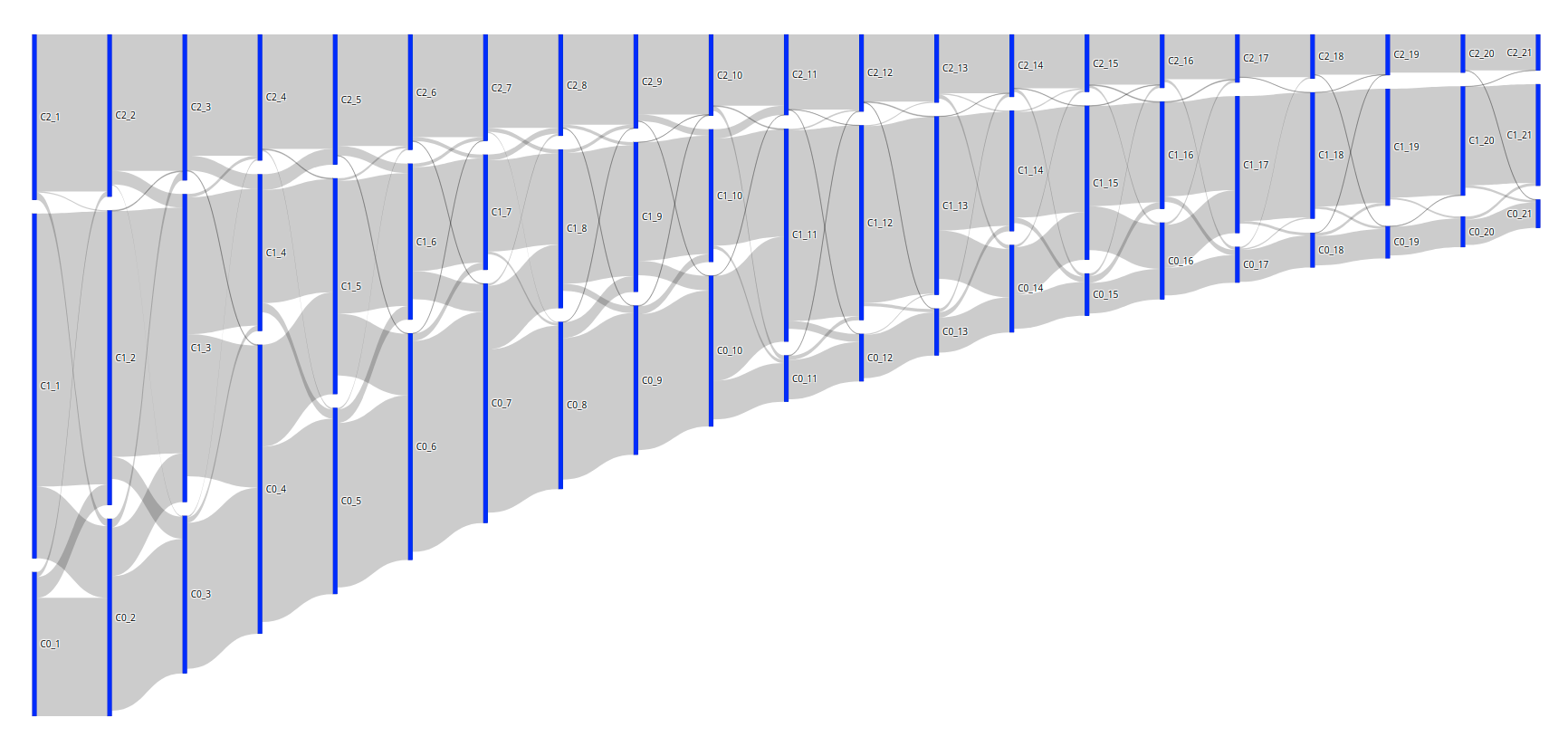
However, I would like to vertically order the bars so that the C0's are always on top, the C1's are always in the middle, and the C2's are always at the bottom (or the other way around, doesn't matter). I know that we can set node.x and node.y to manually assign the coordinates. So, I set the x-values to the amount of days * (1/range of days), which is an increment of +- 0.045. And I set the y-values based on the cluster value: either 0, 0.5 or 1. I then obtain the image below. The vertical order is good, but the vertical margins between the bars are obviously way off; they should be similar to the first result.
The code to produce this is:
import plotly.graph_objects as go
def find_node_coordinates(sources):
x_nodes, y_nodes = [], []
for s in sources:
# Shift each x with +- 0.045
x = float(s.split("_")[-1]) * (1/21)
x_nodes.append(x)
# Choose either 0, 0.5 or 1 for the y-value
cluster_number = s[1]
if cluster_number == "0": y = 1
elif cluster_number == "1": y = 0.5
else: y = 1e-09
y_nodes.append(y)
return x_nodes, y_nodes
def visualize_cluster_flow_counts(counts):
all_sources = list(set(counts['from'].values.tolist() + counts['to'].values.tolist()))
node_x, node_y = find_node_coordinates(all_sources)
froms, tos, vals, labs = [], [], [], []
for index, row in counts.iterrows():
froms.append(all_sources.index(row.values[0]))
tos.append(all_sources.index(row.values[1]))
vals.append(row[2])
labs.append(row[3])
fig = go.Figure(data=[go.Sankey(
arrangement='snap',
node = dict(
pad = 15,
thickness = 5,
line = dict(color = "black", width = 0.1),
label = all_sources,
color = "blue",
x = node_x,
y = node_y,
),
link = dict(
source = froms,
target = tos,
value = vals,
label = labs
))])
fig.update_layout(title_text="Patient flow between clusters over time: 48h (2 days) - 504h (21 days)", font_size=10)
fig.show()
visualize_cluster_flow_counts(counts)
Question: how do I fix the margins of the bars, so that the result looks like the first result? So, for clarity: the bars should be pushed to the bottom. Or is there another way that the Sankey diagram can vertically re-order the bars automatically based on the label value?