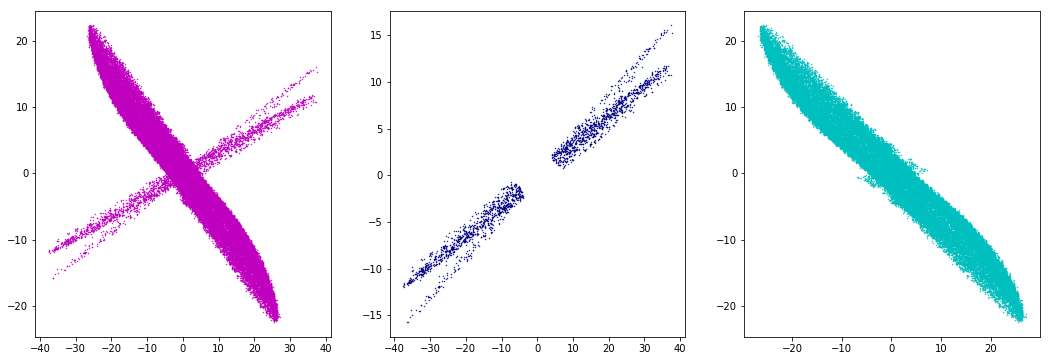
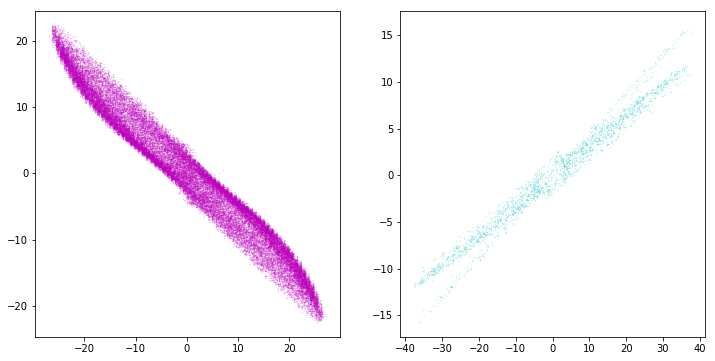I have a data set, which consists of more than one subsets of data. If I plot Y vs. X, I get few overlapping ellipses and I want to cluster them*.
I have tried with the mixture from sklearn, the Bayesian Gaussian Mixture Model gives the best result, however, it does not recognize the overlapping data:
import itertools
import numpy as np
import pylab as plt
from sklearn import mixture
from matplotlib.patches import Ellipse
field_File_1 = './dummy_distrib_3.txt'
'''
link to data:
https://www.dropbox.com/s/jd3wx1ee8r1mj8p/dummy_distrib_3.txt?dl=0
'''
my_dis_1 = np.loadtxt(field_File_1)
X = my_dis_1[:50000,:2]
BaGaMiMo = mixture.BayesianGaussianMixture(n_components=2, covariance_type='full',
weight_concentration_prior_type='dirichlet_distribution').fit(X)
X1 = X[BaGaMiMo.predict(X) == 0, :]
X2 = X[BaGaMiMo.predict(X) == 1, :]
plt.figure(figsize=(18.0, 6.0))
plt.subplot(1,3,1)
plt.scatter(X[:,0], X[:,1], 0.2, color='m')
plt.subplot(1,3,2)
plt.scatter(X[BaGaMiMo.predict(X) == 0, 0], X[BaGaMiMo.predict(X) == 0, 1], .2, color='navy')
plt.subplot(1,3,3)
plt.scatter(X[BaGaMiMo.predict(X) == 1, 0], X[BaGaMiMo.predict(X) == 1, 1], .2, color='c')
plt.show()
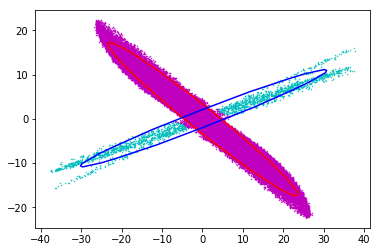
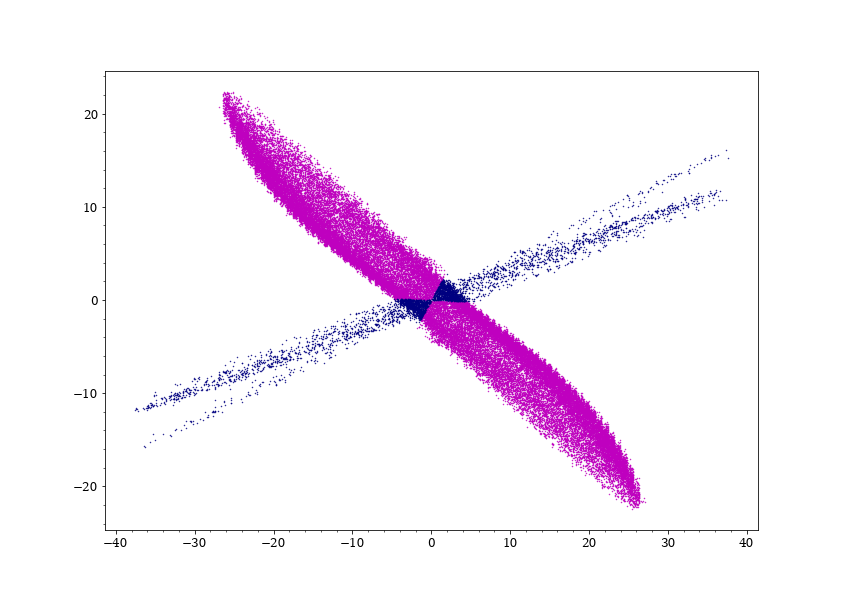
What I do next, is to fit two ellipses to the cyan and navy colored distribution and remove the particles in the cross-section from the cyan distribution,
then assign them randomly to the navy and cyan distributions with the calculated ratio:
One issue is that If I do a histogram of the data, I notice that there is an over-population/discontinuity in the cyan data at the intersection line of the two ellipses and I am looking for ways to reduce that over-population, any help is appreciated.
The jupyter-notebook could be downloaded here: https://www.dropbox.com/s/z1tdgpx1g1lwtb5/Clustering.ipynb?dl=0
.* The data points belong to two sets of charged particles.