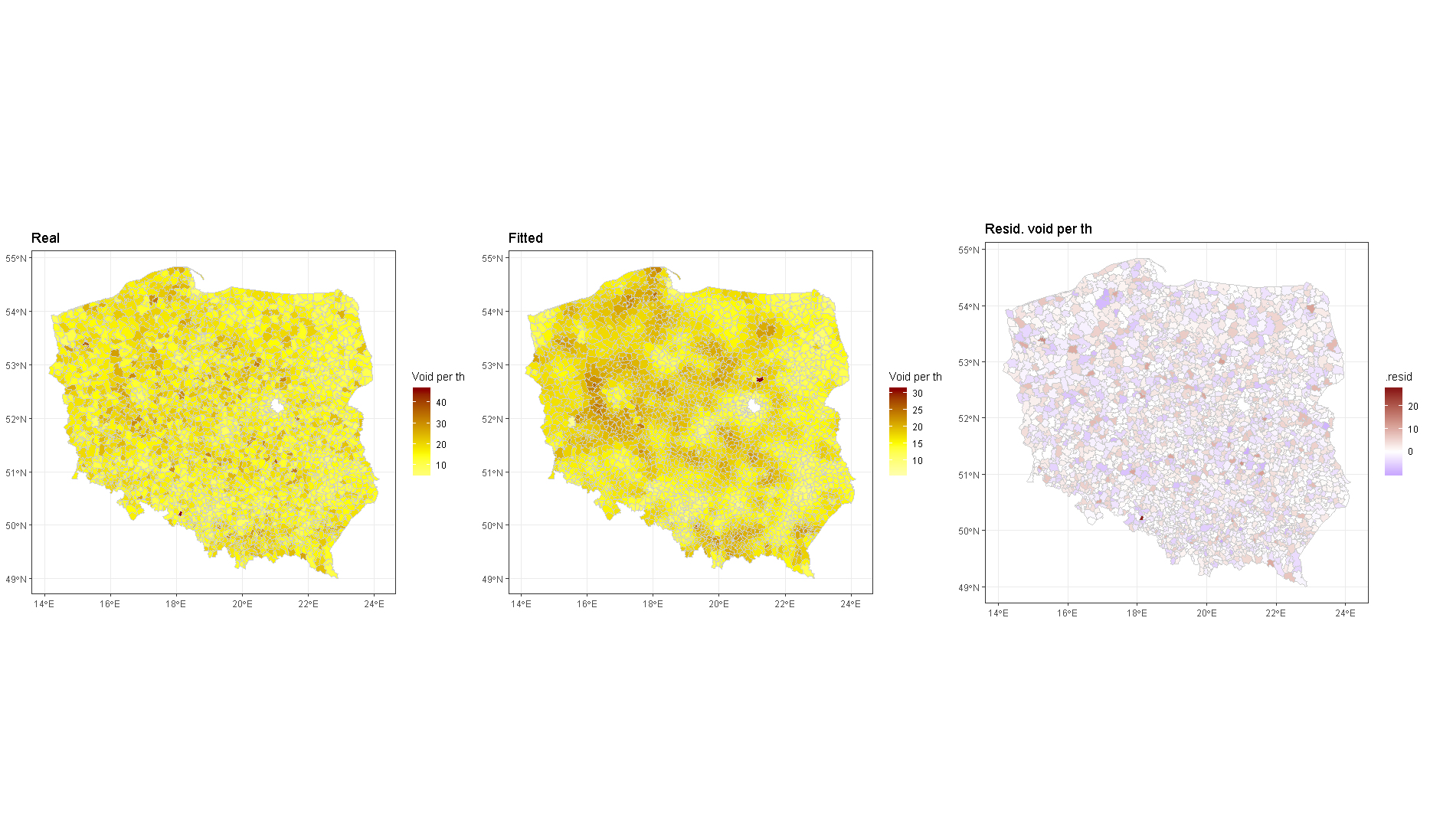
I am trying to fit Polish local government election results in 2015 following the superb blog by @GavinSimpson. https://www.fromthebottomoftheheap.net/2017/10/19/first-steps-with-mrf-smooths/ I joined my xls data with the shp data using a 6 digit identifier (there may be leading 0's). I kept it as a text variable. EDIT, I simplified the identifier and am now using a sequence from 1 to nrow to simplify my question.
library(tidyverse)
library(sf)
library(mgcv)
# Read data
# From https://www.gis-support.pl/downloads/gminy.zip shp file
boroughs_shp <- st_read("../../_mapy/gminy.shp",options = "ENCODING=WINDOWS-1250",
stringsAsFactors = FALSE ) %>%
st_transform(crs = 4326)%>%
janitor::clean_names() %>%
# st_simplify(preserveTopology = T, dTolerance = 0.01) %>%
mutate(teryt=str_sub(jpt_kod_je, 1, 6)) %>%
select(teryt, nazwa=jpt_nazwa, geometry)
# From https://parlament2015.pkw.gov.pl/wyniki_zb/2015-gl-lis-gm.zip data file
elections_xls <-
readxl::read_excel("data/2015-gl-lis-gm.xls",
trim_ws = T, col_names = T) %>%
janitor::clean_names() %>%
select(teryt, liczba_wyborcow, glosy_niewazne)
elections <-
boroughs_shp %>% fortify() %>%
left_join(elections_xls, by = "teryt") %>%
arrange(teryt) %>%
mutate(idx = seq.int(nrow(.)) %>% as.factor(),
teryt = as.factor(teryt))
# Neighbors
boroughs_nb <-spdep::poly2nb(elections, snap = 0.01, queen = F, row.names = elections$idx )
names(boroughs_nb) <- attr(boroughs_nb, "region.id")
# Model
ctrl <- gam.control(nthreads = 4)
m1 <- gam(glosy_niewazne ~ s(idx, bs = 'mrf', xt = list(nb = boroughs_nb)),
data = elections,
offset = log(liczba_wyborcow), # number of votes
method = 'REML',
control = ctrl,
family = betar())
Here is the error message:
Error in smooth.construct.mrf.smooth.spec(object, dk$data, dk$knots) :
mismatch between nb/polys supplied area names and data area names
In addition: Warning message:
In if (all.equal(sort(a.name), sort(levels(k))) != TRUE) stop("mismatch between nb/polys supplied area names and data area names") :
the condition has length > 1 and only the first element will be used
elections$idx is a factor. I am using it to give names to boroughs_nb to be absolutely sure I have the same number of levels. What am I doing wrong?
EDIT: The condition mentioned in error message is met:
> all(sort(names(boroughs_nb)) == sort(levels(elections$idx)))
[1] TRUE

terytis a factor; at least one of the warnings is complain about that. – Bukharins(teryt, bs = "mrf", ...)and the warning implies thatterytis not a factor. Isteryta factor indf_wybory? Does that factor have exactly the same levels as the names of the object you pass tonb? – Bukharinall(sort(names(boroughs_nb)) == sort(levels(idx)))must beTRUEand it isn't; the levels ofidxdon't match the names of the regions in the neighbour list. Basically you need to have a factor with levels that match the names on the neighbour list you pass toxt. – Bukharinall.equal(sort(names(boroughs_nb)), sort(levels(elections$idx)))say? It's failing this check so something isn't right. That's also a bug in mgcv (the warning), but that this is cropping up indicates that mgcv doesn't think these are equal – Bukharin