I'm working on a project where I need to create a preview of nx.Graph() which allows to change position of nodes dragging them with a mouse. My current code is able to redraw whole figure immediately after each motion of mouse if it's clicked on specific node. However, this increases latency significantly. How can I update only artists needed, it is, clicked node, its label text and adjacent edges instead of refreshing every artist of plt.subplots()? Can I at least get a reference to all the artists that need to be relocated?
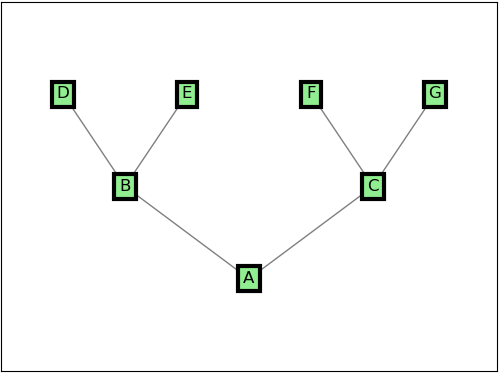
I started from a standard way of displaying a graph in networkx:
import networkx as nx
import matplotlib.pyplot as plt
import numpy as np
import scipy.spatial
def refresh(G):
plt.axis((-4, 4, -1, 3))
nx.draw_networkx_labels(G, pos = nx.get_node_attributes(G, 'pos'),
bbox = dict(fc="lightgreen", ec="black", boxstyle="square", lw=3))
nx.draw_networkx_edges(G, pos = nx.get_node_attributes(G, 'pos'), width=1.0, alpha=0.5)
plt.show()
nodes = np.array(['A', 'B', 'C', 'D', 'E', 'F', 'G'])
edges = np.array([['A', 'B'], ['A', 'C'], ['B', 'D'], ['B', 'E'], ['C', 'F'], ['C', 'G']])
pos = np.array([[0, 0], [-2, 1], [2, 1], [-3, 2], [-1, 2], [1, 2], [3, 2]])
G = nx.Graph()
# IG = InteractiveGraph(G) #>>>>> add this line in the next step
G.add_nodes_from(nodes)
G.add_edges_from(edges)
nx.set_node_attributes(G, dict(zip(G.nodes(), pos.astype(float))), 'pos')
fig, ax = plt.subplots()
# fig.canvas.mpl_connect('button_press_event', lambda event: IG.on_press(event))
# fig.canvas.mpl_connect('motion_notify_event', lambda event: IG.on_motion(event))
# fig.canvas.mpl_connect('button_release_event', lambda event: IG.on_release(event))
refresh(G) # >>>>> replace it with IG.refresh() in the next step
In the next step I changed 5 line of previous script (4 is uncommented and 1 replaced) plus used InteractiveGraph instance to make it interactive:
class InteractiveGraph:
def __init__(self, G, node_pressed=None, xydata=None):
self.G = G
self.node_pressed = node_pressed
self.xydata = xydata
def refresh(self, show=True):
plt.clf()
nx.draw_networkx_labels(self.G, pos = nx.get_node_attributes(self.G, 'pos'),
bbox = dict(fc="lightgreen", ec="black", boxstyle="square", lw=3))
nx.draw_networkx_edges(self.G, pos = nx.get_node_attributes(self.G, 'pos'), width=1.0, alpha=0.5)
plt.axis('off')
plt.axis((-4, 4, -1, 3))
fig.patch.set_facecolor('white')
if show:
plt.show()
def on_press(self, event):
if event.inaxes is not None and len(self.G.nodes()) > 0:
nodelist, coords = zip(*nx.get_node_attributes(self.G, 'pos').items())
kdtree = scipy.spatial.KDTree(coords)
self.xydata = np.array([event.xdata, event.ydata])
close_idx = kdtree.query_ball_point(self.xydata, np.sqrt(0.1))
i = close_idx[0]
self.node_pressed = nodelist[i]
def on_motion(self, event):
if event.inaxes is not None and self.node_pressed:
new_xydata = np.array([event.xdata, event.ydata])
self.xydata += new_xydata - self.xydata
#print(d_xy, self.G.nodes[self.node_pressed])
self.G.nodes[self.node_pressed]['pos'] = self.xydata
self.refresh(show=False)
event.canvas.draw()
def on_release(self, event):
self.node_pressed = None
Related sources:

set_datamethod that you can call to update just that artist, this will allow for significant performance improvements. – Bushelmanipycytoscapeand thanks a lot for reference. I may post a solution if it satisfies my needs. – Arpset_data,set_xyetc.). The relevant part of the code starts here. – Rotherham