For large r and b you can use a method called Monte-Carlo integration, see e.g. Monte Carlo integration on Wikipedia (and/or chapter 3.1.2 of SICP) to compute the sum. For small r and b and significantly different node-failure probabilities p[i] the exact method is superior. The exact definition of small and large will depend on a couple of factors and is best tried out experimentally.
Specific sample code: This is a very basic sample code (in Python) to demonstrate how such a procedure could work:
def montecarlo(p, rb, N):
"""Corresponds to the binomial coefficient formula."""
import random
succ = 0
# Run N samples
for i in xrange(N):
# Generate a single test case
alivenum = 0
for j in xrange(rb):
if random.random()<p: alivenum += 1
# If the test case succeeds, increase succ
if alivenum >= b: succ += 1
# The final result is the number of successful cases/number of total cases
# (I.e., a probability between 0 and 1)
return float(succ)/N
The function corresponds to the binomial test case and runs N tests, checking if b nodes out of r*b nodes are alive with a probability of failure of p. A few experiments will convince you that you need values of N in the range of thousands of samples before you can get any reasonable results, but in principle the complexity is O(N*r*b). The accuracy of the result scales as sqrt(N), i.e., to increase accuracy by a factor of two you need to increase N by a factor of four. For sufficiently large r*b this method will be clearly superior.
Extension of the approximation: You obviously need to design the test case such, that it respects all the properties of the system. You have suggested a couple of extensions, some of which can be easily implemented while others can not. Let me give you a couple of suggestions:
1) In the case of distinct but uncorrelated p[i], the changes of the above code are minimal: In the function head you pass an array instead of a single float p and you replace the line if random.random()<p: alivenum += 1 by
if random.random()<p[j]: alivenum += 1
2) In the case of correlated p[i] you need additional information about the system. The situation I was referring to in my comment could be a network like this:
A--B--C
| |
D E
|
F--G--H
|
J
In this case A might be the "root node" and a failure of node D could imply the automatic failure with 100% probability of nodes F, G, H and J; while a failure of node F would automatically bring down G, H and J etc. At least this was the case I was referring to in my comment (which is a plausible interpretation since you talk about a tree structure of probabilities in the original question). In such a situation you would need modify the code that p refers to a tree structure and for j in ... traverses the tree, skipping the lower branches from the current node as soon as a test fails. The resulting test is still whether alivenum >= b as before, of course.
3) This approach will fail if the network is a cyclic graph that cannot be represented by a tree structure. In such a case you need to first create graph nodes that are either dead or alive and then run a routing algorithm on the graph to count the number of unique, reachable nodes. This won't increase the time-complexity of the algorithm, but obviously the code complexity.
4) Time dependence is a non-trivial, but possible modification if you know the m.t.b.f/r (mean-times-between-failures/repairs) since this can give you the probabilities p of either the tree-structure or the uncorrelated linear p[i] by a sum of exponentials. You will then have to run the MC-procedure at different times with the corresponding results for p.
5) If you merely have the log files (as hinted in your last paragraph) it will require a substantial modification of the approach which is beyond what I can do on this board. The log-files would need to be sufficiently thorough to allow to reconstruct a model for the network graph (and thus the graph of p) as well as the individual values of all nodes of p. Otherwise, accuracy would be unreliable. These log-files would also need to be substantially longer than the time-scales of failures and repairs, an assumptions which may not be realistic in real-life networks.
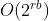 . Also, it doesn't deal with the assumption of node independence.
. Also, it doesn't deal with the assumption of node independence. ?
?
measurement-dateas an additional property. This introduces a timescale into the system, where previously the system was assumed to be static. Previously, a node was either alive (probabilityp) or dead (probability1-p). With a time-scale the system may no longer be static and a certain mean-time-between-failures (for switchingalivenodes todead) and mean-time-between-repairs (the reverse) may become meaningful. If you have this situation, the probability for restoring a file becomes time-dependent. – Ultramicroscopepand1-p, all of which become measurable quantities with a mean value (which may be node-dependent as you have pointed out) and an uncertainty (which reduces to zero if your log becomes long enough). I am pointing this out because in this case the question would be quite different from the original one (where you merely asked for a simplification formula in a specific situation). – Ultramicroscope