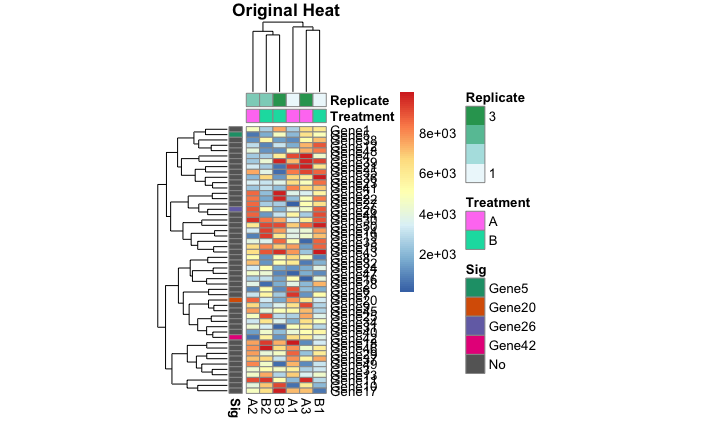
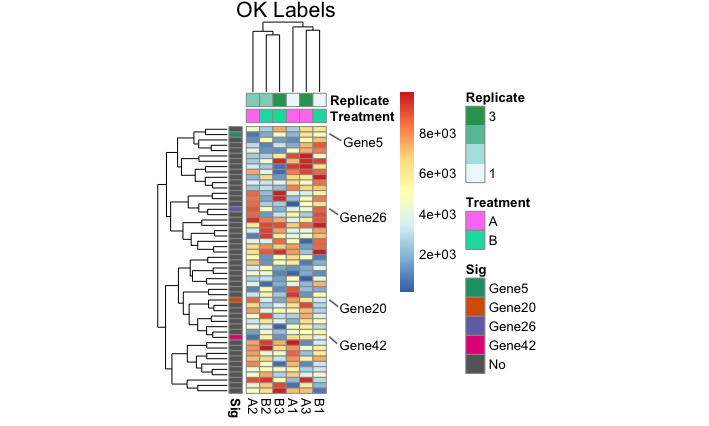
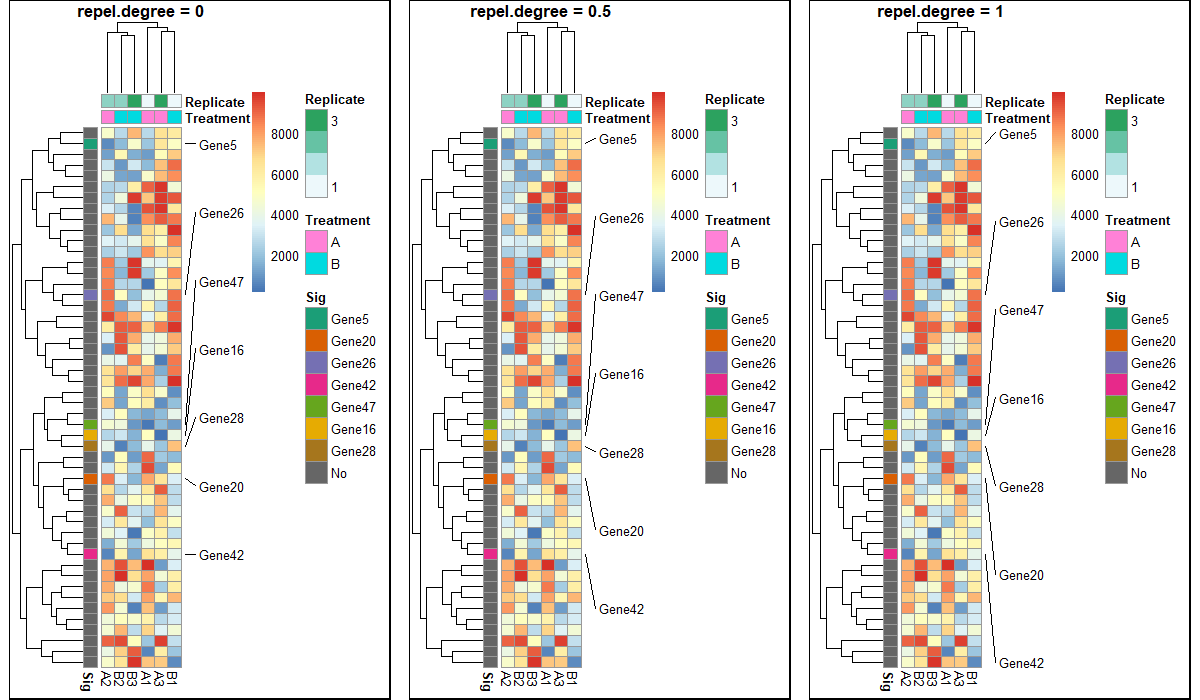
I was wondering if anyone knows of a package that allows partial row labeling of heatmaps. I am currently using pheatmap() to construct my heatmaps, but I can use any package that has this functionality.
I have plots with many rows of differentially expressed genes and I would like to label a subset of them. There are two main things to consider (that I can think of):
- The placement of the text annotation depends on the height of the row. If the rows are too narrow, then the text label will be ambiguous without some sort of pointer.
- If multiple adjacent rows are significant (i.e. will be labelled), then these will need to be offset, and again, a pointer will be needed.
Below is an example of a partial solution that really only gets maybe halfway there, but I hope illustrates what I'd like to be able to do.
set.seed(1)
require(pheatmap)
require(RColorBrewer)
require(grid)
### Data to plot
data_mat <- matrix(sample(1:10000, 300), nrow = 50, ncol = 6)
rownames(data_mat) <- paste0("Gene", 1:50)
colnames(data_mat) <- c(paste0("A", 1:3), paste0("B", 1:3))
### Set how many genes to annotate
### TRUE - make enough labels that some overlap
### FALSE - no overlap
tooMany <- T
### Select a few genes to annotate
if (tooMany) {
sigGenes_v <- paste0("Gene", c(5,20,26,42,47,16,28))
newMain_v <- "Too Many Labels"
} else {
sigGenes_v <- paste0("Gene", c(5,20,26,42))
newMain_v <- "OK Labels"
}
### Make color list
colors_v <- brewer.pal(8, "Dark2")
colors_v <- colors_v[c(1:length(sigGenes_v), 8)]
names(colors_v) <- c(sigGenes_v, "No")
annColors_lsv <- list("Sig" = colors_v)
### Column Metadata
colMeta_df <- data.frame(Treatment = c(rep("A", 3), rep("B", 3)),
Replicate = c(rep(1:3, 2)),
stringsAsFactors = F,
row.names = colnames(data_mat))
### Row metadata
rowMeta_df <- data.frame(Sig = rep("No", 50),
stringsAsFactors = F,
row.names = rownames(data_mat))
for (gene_v in sigGenes_v) rowMeta_df[rownames(rowMeta_df) == gene_v, "Sig"] <- gene_v
### Heatmap
heat <- pheatmap(data_mat,
annotation_row = rowMeta_df,
annotation_col = colMeta_df,
annotation_colors = annColors_lsv,
cellwidth = 10,
main = "Original Heat")
### Get order of genes after clustering
genesInHeatOrder_v <- heat$tree_row$labels[heat$tree_row$order]
whichSigInHeatOrder_v <- which(genesInHeatOrder_v %in% sigGenes_v)
whichSigInHeatOrderLabels_v <- genesInHeatOrder_v[whichSigInHeatOrder_v]
sigY <- 1 - (0.02 * whichSigInHeatOrder_v)
### Change title
whichMainGrob_v <- which(heat$gtable$layout$name == "main")
heat$gtable$grobs[[whichMainGrob_v]] <- textGrob(label = newMain_v,
gp = gpar(fontsize = 16))
### Remove rows
whichRowGrob_v <- which(heat$gtable$layout$name == "row_names")
heat$gtable$grobs[[whichRowGrob_v]] <- textGrob(label = whichSigInHeatOrderLabels_v,
y = sigY,
vjust = 1)
grid.newpage()
grid.draw(heat)
Here are a few outputs:
The "with flags" outputs are the desired final results.
I just saved these as images from the Rstudio plot viewer. I recognize that I could save them as pdfs and provide a larger file size to get rid of the label overlap, but then the individual cells would be larger than I want.