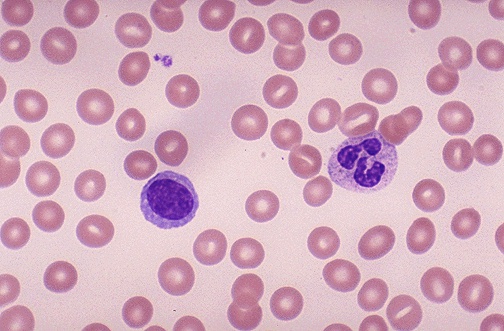
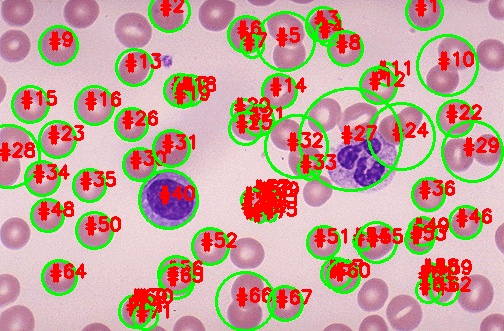
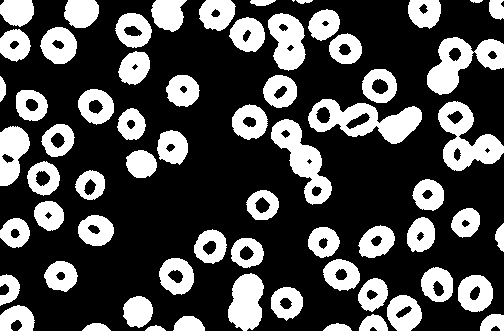
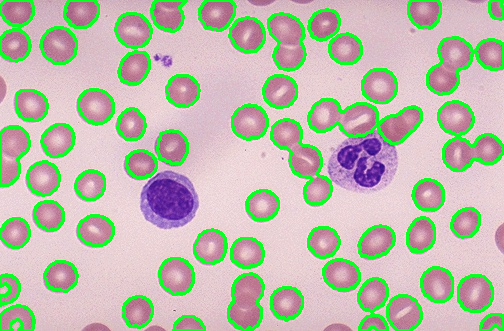
I need code for counting the number of cells in the image and only the cells that are in pink color should be counted .I have used thresholding and watershed method.
import cv2
from skimage.feature import peak_local_max
from skimage.morphology import watershed
from scipy import ndimage
import numpy as np
import imutils
image = cv2.imread("cellorigin.jpg")
gray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
thresh = cv2.threshold(gray, 0, 255,
cv2.THRESH_BINARY_INV | cv2.THRESH_OTSU)[1]
cv2.imshow("Thresh", thresh)
D = ndimage.distance_transform_edt(thresh)
localMax = peak_local_max(D, indices=False, min_distance=20,
labels=thresh)
cv2.imshow("D image", D)
markers = ndimage.label(localMax, structure=np.ones((3, 3)))[0]
labels = watershed(-D, markers, mask=thresh)
print("[INFO] {} unique segments found".format(len(np.unique(labels)) - 1))
for label in np.unique(labels):
# if the label is zero, we are examining the 'background'
# so simply ignore it
if label == 0:
continue
# otherwise, allocate memory for the label region and draw
# it on the mask
mask = np.zeros(gray.shape, dtype="uint8")
mask[labels == label] = 255
# detect contours in the mask and grab the largest one
cnts = cv2.findContours(mask.copy(), cv2.RETR_EXTERNAL,
cv2.CHAIN_APPROX_SIMPLE)
cnts = imutils.grab_contours(cnts)
c = max(cnts, key=cv2.contourArea)
# draw a circle enclosing the object
((x, y), r) = cv2.minEnclosingCircle(c)
cv2.circle(image, (int(x), int(y)), int(r), (0, 255, 0), 2)
cv2.putText(image, "#{}".format(label), (int(x) - 10, int(y)),
cv2.FONT_HERSHEY_SIMPLEX, 0.6, (0, 0, 255), 2)
cv2.imshow("input",image
cv2.waitKey(0)
I am not able to segment the pink cells properly.At some places two pink cells are attached together those also should be separated.
output: