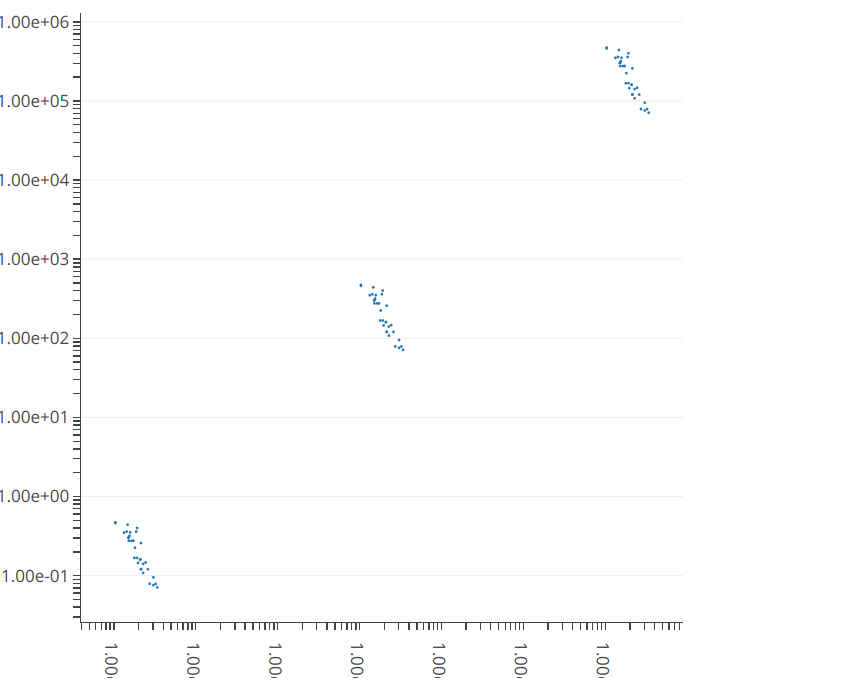
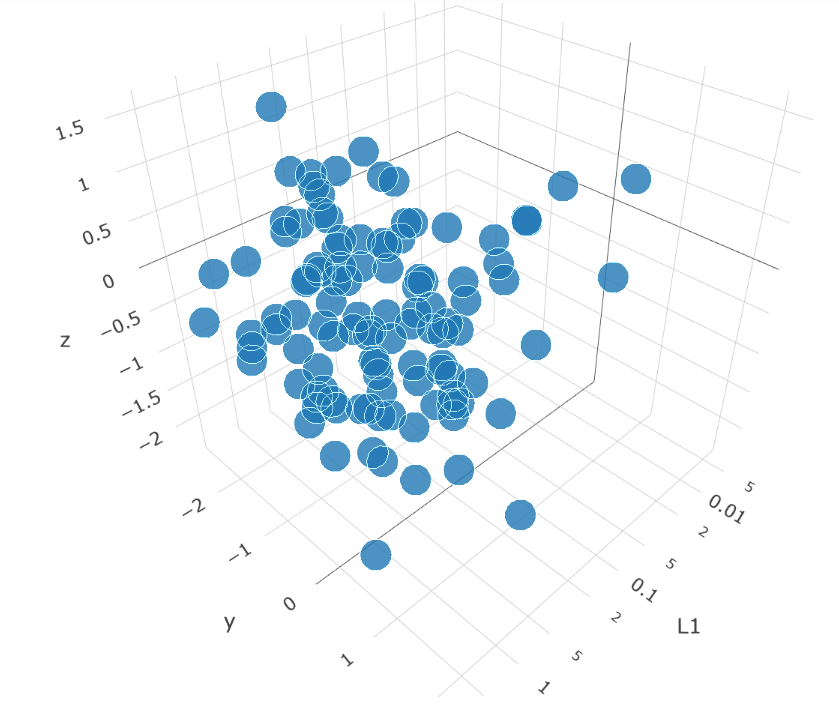
I've build the following test app where I solve the issue to get the tick labels as scientific annotation, but I would now like to reduce the number of grid lines to only be placed at the "main" ticks, i.e. the ones that have a text label. This question was posted based on discussion / comment on this previous SO question
I would like to find a way that works for both 2D and 3D plotly scatter plots since I am using both.
Here is the 3D app.
library(shiny)
library(plotly)
shinyApp(
ui = fluidPage( plotlyOutput('plot') ),
server = function(input, output) {
output$plot <- renderPlotly ({
mtcars <- rbind(mtcars, mtcars*1000, mtcars/1000) #create data with big logarithmic range
maxlog <- round(log10(max(mtcars[['mpg']][mtcars[['mpg']]>0], mtcars[['disp']][mtcars[['disp']]>0],mtcars[['cyl']][mtcars[['cyl']]>0])), digits = 0) +1 # determine max log needed
minlog <- round(log10(min(mtcars[['mpg']][mtcars[['mpg']]>0], mtcars[['disp']][mtcars[['disp']]>0],mtcars[['cyl']][mtcars[['cyl']]>0])), digits = 0) -1 # determine min log needed
logrange <- (maxlog - minlog)*9 +1 # get the distance between smallest and largest log power
tval <- sort(as.vector(sapply(seq(1,9), function(x) x*10^seq(minlog, maxlog)))) #generates a sequence of numbers in logarithmic divisions
ttxt <- rep("",length(tval)) # no label at most of the ticks
ttxt[seq(1,logrange,9)] <- formatC(tval, format = "e", digits = 2)[seq(1,logrange,9)] # every 9th tick is labelled
p <- plot_ly(source = 'ThresholdScatter')
p <- add_trace(p, data = mtcars,
x = mtcars[['mpg']],
y = mtcars[['disp']],
z = mtcars[['cyl']],
type = 'scatter3d',
mode = 'markers',
marker = list(size = 2))
p <- layout(p, autosize = F, width = 500, height = 500,
scene = list(yaxis = list(type="log",
zeroline=F, showline=T,
ticks="outside",
tickvals=tval,
ticktext=ttxt),
xaxis = list(type="log",
zeroline=F, showline=T,
ticks="outside",
tickvals=tval,
ticktext=ttxt),
zaxis = list(type="log",
zeroline=F, showline=T,
ticks="outside",
tickvals=tval,
ticktext=ttxt),
camera = list(eye = list(x = -1.5, y = 1.5, z = 1.5))))
})
}
)
and the same but in 2D
library(shiny)
library(plotly)
shinyApp(
ui = fluidPage( plotlyOutput('plot') ),
server = function(input, output) {
output$plot <- renderPlotly ({
mtcars <- rbind(mtcars, mtcars*1000, mtcars/1000) #create data with big logarithmic range
maxlog <- round(log10(max(mtcars[['mpg']][mtcars[['mpg']]>0], mtcars[['disp']][mtcars[['disp']]>0])), digits = 0) +1 # determine max log needed
minlog <- round(log10(min(mtcars[['mpg']][mtcars[['mpg']]>0], mtcars[['disp']][mtcars[['disp']]>0])), digits = 0) -1 # determine min log needed
logrange <- (maxlog - minlog)*9 +1 # get the distance between smallest and largest log power
tval <- sort(as.vector(sapply(seq(1,9), function(x) x*10^seq(minlog,
maxlog)))) #generates a sequence of numbers in logarithmic divisions
ttxt <- rep("",length(tval)) # no label at most of the ticks
ttxt[seq(1,logrange,9)] <- formatC(tval, format = "e", digits = 2)[seq(1,logrange,9)] # every 9th tick is labelled
p <- plot_ly(source = 'ThresholdScatter')
p <- add_trace(p, data = mtcars,
x = mtcars[['mpg']],
y = mtcars[['disp']],
type = 'scatter',
mode = 'markers',
marker = list(size = 2))
p <- layout(p,autosize = F, width = 500, height = 500,
yaxis = list(type="log",
zeroline=F, showline=T,
ticks="outside",
tickvals=tval,
ticktext=ttxt),
xaxis = list(type="log",
zeroline=F, showline=T,
ticks="outside",
tickvals=tval,
ticktext=ttxt))
})
}
)