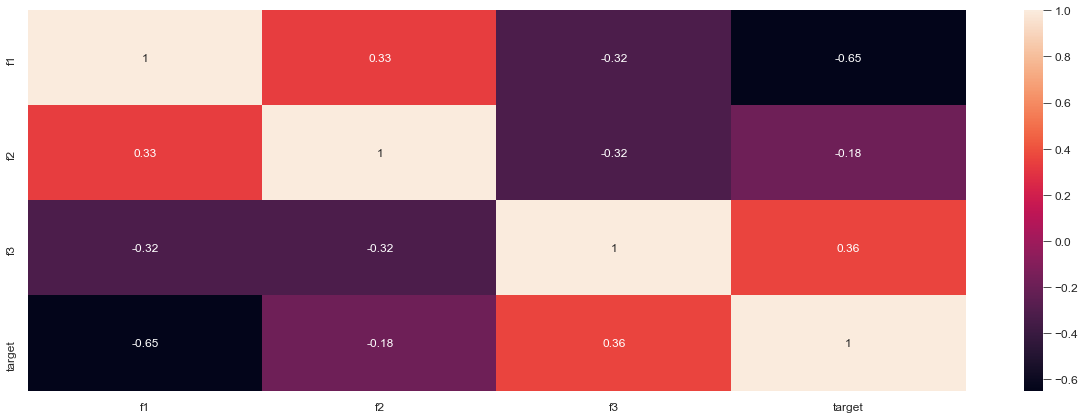
Step 0: Problem description
I have a classification problem, ie I want to predict a binary target based on a collection of numerical features, using logistic regression, and after running a Principal Components Analysis (PCA).
I have 2 datasets: df_train and df_valid (training set and validation set respectively) as pandas data frame, containing the features and the target. As a first step, I have used get_dummies pandas function to transform all the categorical variables as boolean. For example, I would have:
n_train = 10
np.random.seed(0)
df_train = pd.DataFrame({"f1":np.random.random(n_train), \
"f2": np.random.random(n_train), \
"f3":np.random.randint(0,2,n_train).astype(bool),\
"target":np.random.randint(0,2,n_train).astype(bool)})
In [36]: df_train
Out[36]:
f1 f2 f3 target
0 0.548814 0.791725 False False
1 0.715189 0.528895 True True
2 0.602763 0.568045 False True
3 0.544883 0.925597 True True
4 0.423655 0.071036 True True
5 0.645894 0.087129 True False
6 0.437587 0.020218 True True
7 0.891773 0.832620 True False
8 0.963663 0.778157 False False
9 0.383442 0.870012 True True
n_valid = 3
np.random.seed(1)
df_valid = pd.DataFrame({"f1":np.random.random(n_valid), \
"f2": np.random.random(n_valid), \
"f3":np.random.randint(0,2,n_valid).astype(bool),\
"target":np.random.randint(0,2,n_valid).astype(bool)})
In [44]: df_valid
Out[44]:
f1 f2 f3 target
0 0.417022 0.302333 False False
1 0.720324 0.146756 True False
2 0.000114 0.092339 True True
I would like now to apply a PCA to reduce the dimensionality of my problem, then use LogisticRegression from sklearn to train and get prediction on my validation set, but I'm not sure the procedure I follow is correct. Here is what I do:
Step 1: PCA
The idea is that I need to transform both my training and validation set the same way with PCA. In other words, I can not perform PCA separately. Otherwise, they will be projected on different eigenvectors.
from sklearn.decomposition import PCA
pca = PCA(n_components=2) #assume to keep 2 components, but doesn't matter
newdf_train = pca.fit_transform(df_train.drop("target", axis=1))
newdf_valid = pca.transform(df_valid.drop("target", axis=1)) #not sure here if this is right
Step2: Logistic Regression
It's not necessary, but I prefer to keep things as dataframe:
features_train = pd.DataFrame(newdf_train)
features_valid = pd.DataFrame(newdf_valid)
And now I perform the logistic regression
from sklearn.linear_model import LogisticRegression
cls = LogisticRegression()
cls.fit(features_train, df_train["target"])
predictions = cls.predict(features_valid)
I think step 2 is correct, but I have more doubts about step 1: is this the way I'm supposed to chain PCA, then a classifier ?