I am trying to learn how to sample truncated distributions. To begin with I decided to try a simple example I found here example
I didn't really understand the division by the CDF, therefore I decided to tweak the algorithm a bit. Being sampled is an exponential distribution for values x>0 Here is an example python code:
# Sample exponential distribution for the case x>0
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import norm
def pdf(x):
return x*np.exp(-x)
xvec=np.zeros(1000000)
x=1.
for i in range(1000000):
a=x+np.random.normal()
xs=x
if a > 0. :
xs=a
A=pdf(xs)/pdf(x)
if np.random.uniform()<A :
x=xs
xvec[i]=x
x=np.linspace(0,15,1000)
plt.plot(x,pdf(x))
plt.hist([x for x in xvec if x != 0],bins=150,normed=True)
plt.show()
The code above seems to work fine only for when using the condition if a > 0. :, i.e. positive x, choosing another condition (e.g. if a > 0.5 :) produces wrong results.
Since my final goal was to sample a 2D-Gaussian - pdf on a truncated interval I tried extending the simple example using the exponential distribution (see the code below). Unfortunately, since the simple case didn't work, I assume that the code given below would yield wrong results.
I assume that all this can be done using the advanced tools of python. However, since my primary idea was to understand the principle behind, I would greatly appreciate your help to understand my mistake. Thank you for your help.
EDIT:
# code updated according to the answer of CrazyIvan
from scipy.stats import multivariate_normal
RANGE=100000
a=2.06072E-02
b=1.10011E+00
a_range=[0.001,0.5]
b_range=[0.01, 2.5]
cov=[[3.1313994E-05, 1.8013737E-03],[ 1.8013737E-03, 1.0421529E-01]]
x=a
y=b
j=0
for i in range(RANGE):
a_t,b_t=np.random.multivariate_normal([a,b],cov)
# accept if within bounds - all that is neded to truncate
if a_range[0]<a_t and a_t<a_range[1] and b_range[0]<b_t and b_t<b_range[1]:
print(dx,dy)
EDIT:
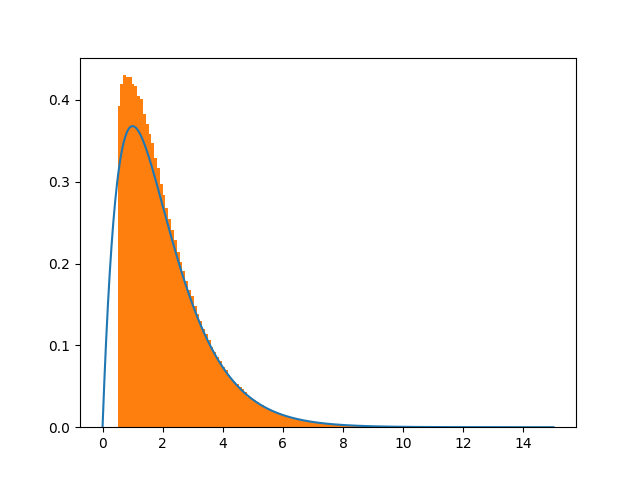
I changed the code by norming the analytic pdf according to this scheme, and according to the answers given by, @Crazy Ivan and @Leandro Caniglia , for the case where the bottom of the pdf is removed. That is dividing by (1-CDF(0.5)) since my accept condition is x>0.5. This seems again to show some discrepancies. Again the mystery prevails ..
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import norm
def pdf(x):
return x*np.exp(-x)
# included the corresponding cdf
def cdf(x):
return 1. -np.exp(-x)-x*np.exp(-x)
xvec=np.zeros(1000000)
x=1.
for i in range(1000000):
a=x+np.random.normal()
xs=x
if a > 0.5 :
xs=a
A=pdf(xs)/pdf(x)
if np.random.uniform()<A :
x=xs
xvec[i]=x
x=np.linspace(0,15,1000)
# new part norm the analytic pdf to fix the area
plt.plot(x,pdf(x)/(1.-cdf(0.5)))
plt.hist([x for x in xvec if x != 0],bins=200,normed=True)
plt.savefig("test_exp.png")
plt.show()
It seems that this can be cured by choosing larger shift size
shift=15.
a=x+np.random.normal()*shift.
which is in general an issue of the Metropolis - Hastings. See the graph below:
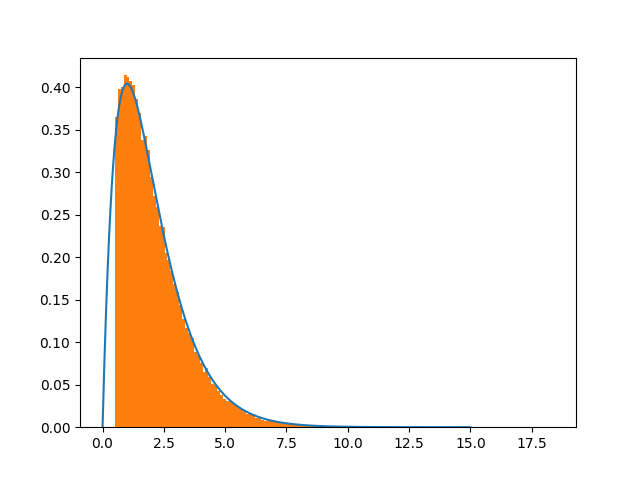
Bottom line is that changing the shift size definitely improves the convergence. The misery is why, since the Gaussian is unbounded.


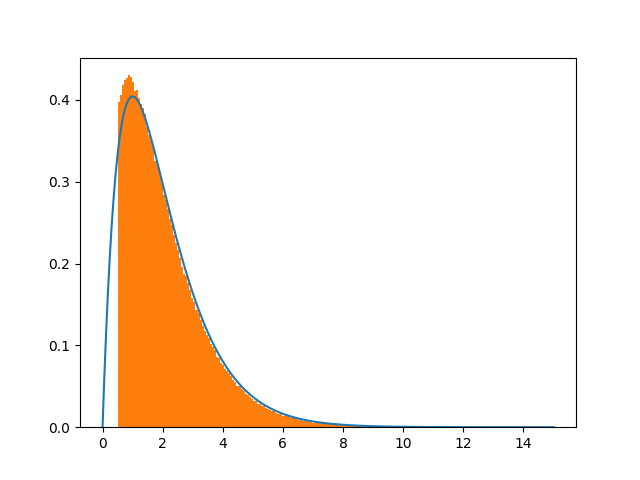
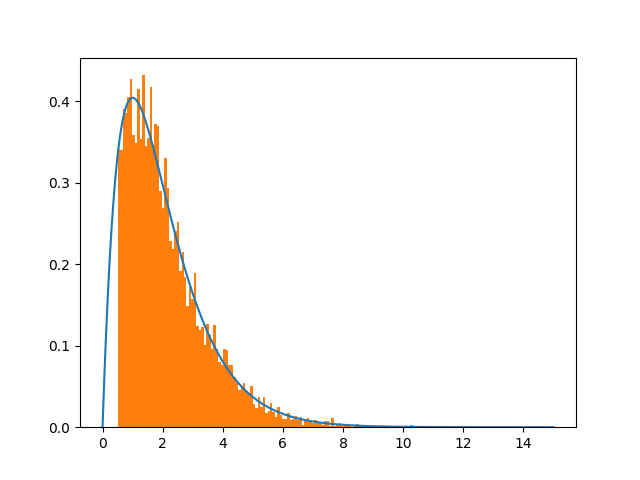

a > 0.5? The excess area of the orange part seems to compensate the part below the curve withx<0.5, which is what you want so that the total orange area (i.e., the integral of the truncatedpdf)equals1. – Duckpdf(x)/(1-cdf(0.5))in your case. This is precisely what my answer says (see below) because in your caseB=1andA=cdf(0.5). I've rewritten this part of the answer to make it clearer. – Duck0.5<x<1.2Of course to properly plot, I have to cut the unnecessary part from the blue curve. – Binetta