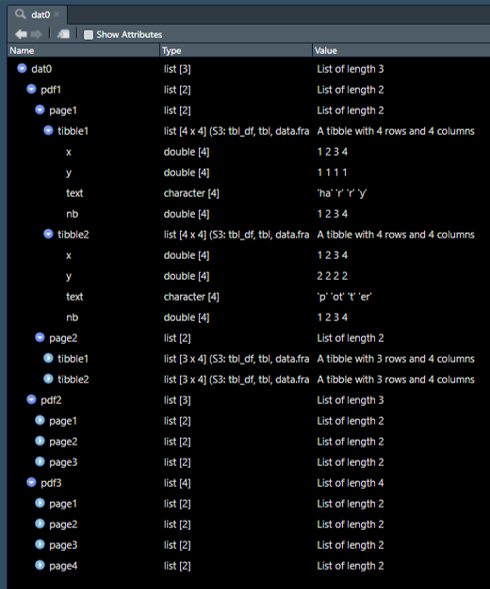
Beginner in data manipulation in R, I struggle with multi-level nested lists.
Question: Is there a way to transform this dat0 3-levels list into the global dat1 dataframe below?
- The new
fulltextcolumn concatenates thetextvariable from each tibble. - the new
nbsumcolumn adds thenbvariable from each tibble.
Note: a purrr-based approach with dplyr functions (mutate...) would be welcome to better understand these tools in this particular context. Other approaches are also welcome!
Thanks for help
Initial data:
dat0 <- list(pdf1 =
list(page1 =
list(tibble1 = tibble(x = c(1,2,3,4), y = c(1,1,1,1), text = c("ha","r","r","y"), nb = c(1,2,3,4)),
tibble2 = tibble(x = c(1,2,3,4), y = c(2,2,2,2), text = c("p","ot","t","er"), nb = c(1,2,3,4))),
page2 =
list(tibble1 = tibble(x = c(1,2,3), y = c(3,3,3), text = c("her","m","ione"), nb = c(1,2,3)),
tibble2 = tibble(x = c(1,2,3), y = c(4,4,4), text = c("gra","ng","er"), nb = c(1,2,3)))),
pdf2 =
list(page1 =
list(tibble1 = tibble(x = c(1,2), y = c(5,5), text = c("vol","de"), nb = c(1,2)),
tibble2 = tibble(x = c(1,2), y = c(6,6), text = c("m","ort"), nb = c(1,2))),
page2 =
list(tibble1 = tibble(x = c(1,2,3,4,5), y = c(7,7,7,7,7), text = c("a","l","b","u","s"), nb = c(1,2,3,4,5)),
tibble2 = tibble(x = c(1,2,3,4,5), y = c(8,8,8,8,8), text = c("du","m","ble","do","re"), nb = c(1,2,3,4,5))),
page3 =
list(tibble1 = tibble(x = c(1,2,3,4), y = c(9,9,9,9), text = c("dr","a","g","o"), nb = c(1,2,3,4)),
tibble2 = tibble(x = c(1,2,3,4), y = c(10,10,10,10), text = c("ma","lf","o","y"), nb = c(1,2,3,4)))),
pdf3 =
list(page1 =
list(tibble1 = tibble(x = c(1,2,3,4,5), y = c(11,11,11,11,11), text = c("s","ev","e","ru","s"), nb = c(1,2,3,4,5)),
tibble2 = tibble(x = c(1,2,3,4,5), y = c(12,12,12,12,12), text = c("r","o","g","u","e"), nb = c(1,2,3,4,5))),
page2 =
list(tibble1 = tibble(x = c(1,2,3), y = c(13,13,13), text = c("r","o","n"), nb = c(1,2,3)),
tibble2 = tibble(x = c(1,2,3), y = c(14,14,14), text = c("we","as","ley"), nb = c(1,2,3))),
page3 =
list(tibble1 = tibble(x = c(1,2,3,4,5,6), y = c(15,15,15,15,15,15), text = c("be","l","la","t","ri","x"), nb = c(1,2,3,4,5,6)),
tibble2 = tibble(x = c(1,2,3,4,5,6), y = c(16,16,16,16,16,16), text = c("l","est","r","a","ng","e"), nb = c(1,2,3,4,5,6))),
page4 =
list(tibble1 = tibble(x = c(1,2), y = c(17,17), text = c("sir","ius"), nb = c(1,2)),
tibble2 = tibble(x = c(1,2), y = c(18,18), text = c("bl","ack"), nb = c(1,2)))))
Desired output (constructed laboriously; dput script below):
dat1 <-
structure(list(pdf = c("pdf1", "pdf1", "pdf1", "pdf1", "pdf1",
"pdf1", "pdf1", "pdf1", "pdf1", "pdf1", "pdf1", "pdf1", "pdf1",
"pdf1", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2",
"pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2",
"pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf2", "pdf3",
"pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3",
"pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3",
"pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3",
"pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3", "pdf3"), page = c("page1",
"page1", "page1", "page1", "page1", "page1", "page1", "page1",
"page2", "page2", "page2", "page2", "page2", "page2", "page1",
"page1", "page1", "page1", "page2", "page2", "page2", "page2",
"page2", "page2", "page2", "page2", "page2", "page2", "page3",
"page3", "page3", "page3", "page3", "page3", "page3", "page3",
"page1", "page1", "page1", "page1", "page1", "page1", "page1",
"page1", "page1", "page1", "page2", "page2", "page2", "page2",
"page2", "page2", "page3", "page3", "page3", "page3", "page3",
"page3", "page3", "page3", "page3", "page3", "page3", "page3",
"page4", "page4", "page4", "page4"), x = c(1, 2, 3, 4, 1, 2,
3, 4, 1, 2, 3, 1, 2, 3, 1, 2, 1, 2, 1, 2, 3, 4, 5, 1, 2, 3, 4,
5, 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4, 5, 1, 2, 3, 4, 5, 1, 2,
3, 1, 2, 3, 1, 2, 3, 4, 5, 6, 1, 2, 3, 4, 5, 6, 1, 2, 1, 2),
y = c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 4, 4, 4, 5, 5, 6,
6, 7, 7, 7, 7, 7, 8, 8, 8, 8, 8, 9, 9, 9, 9, 10, 10, 10,
10, 11, 11, 11, 11, 11, 12, 12, 12, 12, 12, 13, 13, 13, 14,
14, 14, 15, 15, 15, 15, 15, 15, 16, 16, 16, 16, 16, 16, 17,
17, 18, 18), text = c("ha", "r", "r", "y", "p", "ot", "t",
"er", "her", "m", "ione", "gra", "ng", "er", "vol", "de",
"m", "ort", "a", "l", "b", "u", "s", "du", "m", "ble", "do",
"re", "dr", "a", "g", "o", "ma", "lf", "o", "y", "s", "ev",
"e", "ru", "s", "r", "o", "g", "u", "e", "r", "o", "n", "we",
"as", "ley", "be", "l", "la", "t", "ri", "x", "l", "est",
"r", "a", "ng", "e", "sir", "ius", "bl", "ack"), nb = c(1,
2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 1, 2, 3, 1, 2, 1, 2, 1, 2,
3, 4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3,
4, 5, 1, 2, 3, 4, 5, 1, 2, 3, 1, 2, 3, 1, 2, 3, 4, 5, 6,
1, 2, 3, 4, 5, 6, 1, 2, 1, 2), fulltext = c("harry", "harry",
"harry", "harry", "potter", "potter", "potter", "potter",
"hermione", "hermione", "hermione", "granger", "granger",
"granger", "volde", "volde", "mort", "mort", "albus", "albus",
"albus", "albus", "albus", "dumbledore", "dumbledore", "dumbledore",
"dumbledore", "dumbledore", "drago", "drago", "drago", "drago",
"malfoy", "malfoy", "malfoy", "malfoy", "severus", "severus",
"severus", "severus", "severus", "rogue", "rogue", "rogue",
"rogue", "rogue", "ron", "ron", "ron", "weasley", "weasley",
"weasley", "bellatrix", "bellatrix", "bellatrix", "bellatrix",
"bellatrix", "bellatrix", "lestrange", "lestrange", "lestrange",
"lestrange", "lestrange", "lestrange", "sirius", "sirius",
"black", "black"), nbsum = c(10, 10, 10, 10, 10, 10, 10,
10, 6, 6, 6, 6, 6, 6, 3, 3, 3, 3, 15, 15, 15, 15, 15, 15,
15, 15, 15, 15, 10, 10, 10, 10, 10, 10, 10, 10, 15, 15, 15,
15, 15, 15, 15, 15, 15, 15, 6, 6, 6, 6, 6, 6, 21, 21, 21,
21, 21, 21, 21, 21, 21, 21, 21, 21, 3, 3, 3, 3)), row.names = c(NA,
-68L), class = "data.frame")