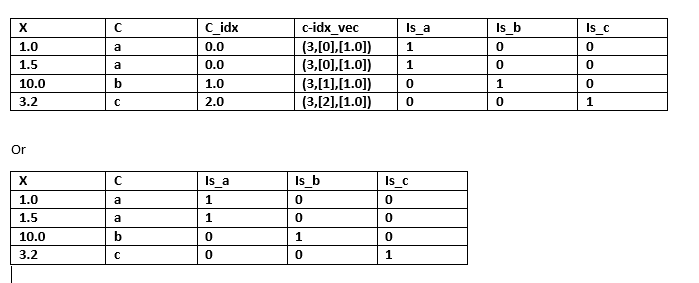
I am using apache Spark ML lib to handle categorical features using one hot encoding. After writing the below code I am getting a vector c_idx_vec as output of one hot encoding. I do understand how to interpret this output vector but I am unable to figure out how to convert this vector into columns so that I get a new transformed dataframe.Take this dataset for example:
>>> fd = spark.createDataFrame( [(1.0, "a"), (1.5, "a"), (10.0, "b"), (3.2, "c")], ["x","c"])
>>> ss = StringIndexer(inputCol="c",outputCol="c_idx")
>>> ff = ss.fit(fd).transform(fd)
>>> ff.show()
+----+---+-----+
| x| c|c_idx|
+----+---+-----+
| 1.0| a| 0.0|
| 1.5| a| 0.0|
|10.0| b| 1.0|
| 3.2| c| 2.0|
+----+---+-----+
By default, the OneHotEncoder will drop the last category:
>>> oe = OneHotEncoder(inputCol="c_idx",outputCol="c_idx_vec")
>>> fe = oe.transform(ff)
>>> fe.show()
+----+---+-----+-------------+
| x| c|c_idx| c_idx_vec|
+----+---+-----+-------------+
| 1.0| a| 0.0|(2,[0],[1.0])|
| 1.5| a| 0.0|(2,[0],[1.0])|
|10.0| b| 1.0|(2,[1],[1.0])|
| 3.2| c| 2.0| (2,[],[])|
+----+---+-----+-------------+
Of course, this behavior can be changed:
>>> oe.setDropLast(False)
>>> fl = oe.transform(ff)
>>> fl.show()
+----+---+-----+-------------+
| x| c|c_idx| c_idx_vec|
+----+---+-----+-------------+
| 1.0| a| 0.0|(3,[0],[1.0])|
| 1.5| a| 0.0|(3,[0],[1.0])|
|10.0| b| 1.0|(3,[1],[1.0])|
| 3.2| c| 2.0|(3,[2],[1.0])|
+----+---+-----+-------------+
So, I wanted to know how to convert my c_idx_vec vector into new dataframe as below: