You can try this approach:
f <- function(n=5,m=3)
t(apply(combn(1:n,m=m),2,function(cm) replace(rep(0,n),cm,1)))
f(5,3)
# [,1] [,2] [,3] [,4] [,5]
# [1,] 1 1 1 0 0
# [2,] 1 1 0 1 0
# [3,] 1 1 0 0 1
# [4,] 1 0 1 1 0
# [5,] 1 0 1 0 1
# [6,] 1 0 0 1 1
# [7,] 0 1 1 1 0
# [8,] 0 1 1 0 1
# [9,] 0 1 0 1 1
# [10,] 0 0 1 1 1
The idea is to generate all combinations of indices for 1, and then to use them to produce the final result.
Another flavor of the same approach:
f.2 <- function(n=5,m=3)
t(combn(1:n,m,FUN=function(cm) replace(rep(0,n),cm,1)))
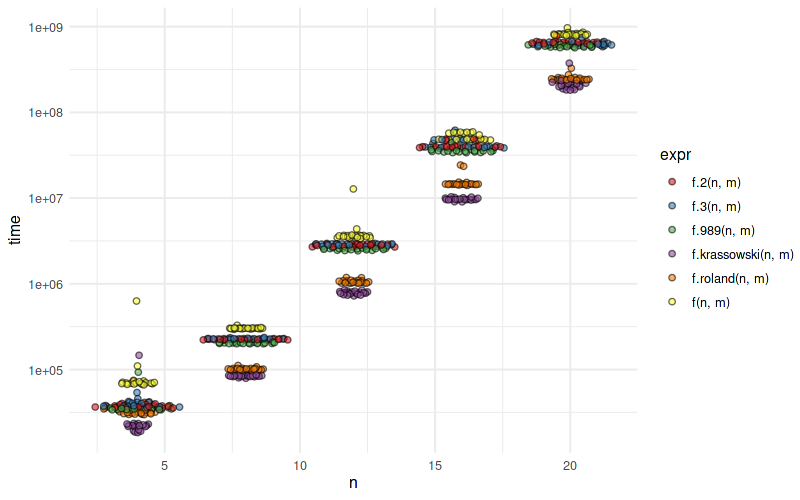
The second approach is about twice faster:
library(rbenchmark)
benchmark(f(16,8),f.2(16,8))
# test replications elapsed relative user.self sys.self user.child sys.child
# 2 f.2(16, 8) 100 5.706 1.000 5.688 0.017 0 0
# 1 f(16, 8) 100 10.802 1.893 10.715 0.082 0 0
Benchmark
f.akrun <- function(n=5,m=3) {
indx <- combnPrim(1:n,m)
DT <- setDT(as.data.frame(matrix(0, ncol(indx),n)))
for(i in seq_len(nrow(DT))){
set(DT, i=i, j=indx[,i],value=1)
}
DT
}
benchmark(f(16,8),f.2(16,8),f.akrun(16,8))
# test replications elapsed relative user.self sys.self user.child sys.child
# 2 f.2(16, 8) 100 5.464 1.097 5.435 0.028 0 0
# 3 f.akrun(16, 8) 100 4.979 1.000 4.938 0.037 0 0
# 1 f(16, 8) 100 10.854 2.180 10.689 0.129 0 0
@akrun's solution (f.akrun) is ~10% faster than f.2.
[EDIT]
Another approach, which is even more faster and simple:
f.3 <- function(n=5,m=3) t(combn(n,m,tabulate,nbins=n))

x <- expand.grid(rep(list(0L:1L), 5L)); x[rowSums(x) ==3L,]but I think you want something faster than that. – Slobber