You could write your own function to do this. It takes three arguments: the number of points you want, the radius of the containing circle, and the minimum distance between points.
It simply starts with two empty vectors for x and y, then generates a random x, y pair drawn from the uniform distribution. If this x, y pair is outside the unit circle or within a given distance of an existing point, it is discarded and another pair drawn. Otherwise the point is kept. This process is repeated until the x and y vectors are full. At this point, a data frame is created of the x and y values multiplied by the desired circle radius to generate the result.
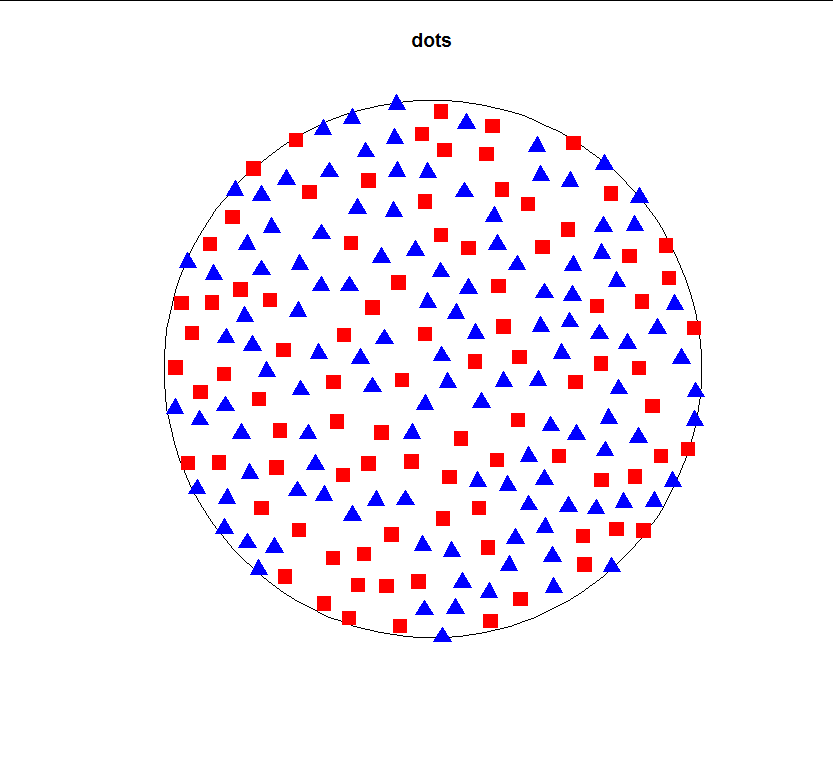
If it cannot find any places to put a new point after trying a large number of times it will throw an error. The given example of 210 points will only just fit if the minimum distance is 0.1:
repelled_points <- function(n, r_circle, r_clash) {
container_x <- numeric(n)
container_y <- numeric(n)
j <- i <- 1
while(i <= n)
{
j <- j + 1
if(j == 100 * n) stop("Cannot accommodate the points in given space")
x <- runif(1, -1, 1)
y <- runif(1, -1, 1)
if(x^2 + y^2 > 1) next
if(i > 1) {
dist <- sqrt((x - container_x[seq(i-1)])^2 + (y - container_y[seq(i-1)])^2)
if(any(dist < r_clash)) next
}
container_x[i] <- x
container_y[i] <- y
i <- i + 1
j <- 1
}
`class<-`(list(window = disc(centre = c(0, 0), radius = r_circle),
n = n, x = container_x * r_circle,
y = container_y * r_circle, markformat = "none"), "ppp")
}
Which, when you run your plotting code, returns the following result:
dots <- repelled_points(210, 1, 0.1)
plot(dots, type="n")
points(dots$x[1:45], dots$y[1:45], pch=15, col="red", cex=2)
points(dots$x[46:90], dots$y[46:90], pch=15, col="red", cex=2)
points(dots$x[91:151], dots$y[91:151], pch=17, col="blue", cex=2)
points(dots$x[152:210], dots$y[152:210], pch=17, col="blue", cex=2)
![enter image description here]()



spatial::Straussdoes something like this, although it looks like it only does it in a rectangular domain, so you'd first have to generate the Strauss process on the rectangular domain then subset it to the circular domain. – Guaiacol