I'm working in RStudio on a simple analysis where I source some files via the source command. For example, I have this file with some simple analysis:
analysis.R
# Settings ----------------------------------------------------------------
data("mtcars")
source("Generic Functions.R")
# Some work ---------------------------------------------------------------
# Makes no sense
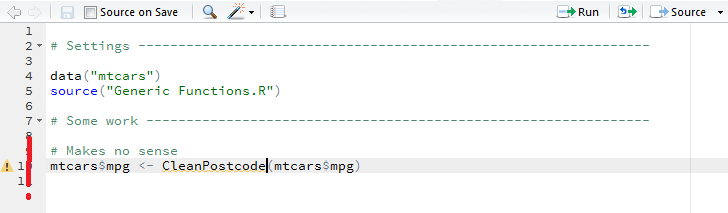
mtcars$mpg <- CleanPostcode(mtcars$mpg)
The generic functions file has some simple functions that I use to derive graphs and do repetitive tasks. For example the used CleanPostcode function would look like that:
Generic Functions.R
#' The file provides a set of generic functions
# String manipulations ----------------------------------------------------
# Create a clean Postcode for matching
CleanPostcode <- function(MessyPostcode) {
MessyPostcode <- as.character(MessyPostcode)
MessyPostcode <- gsub("[[:space:]]", "", MessyPostcode)
MessyPostcode <- gsub("[[:punct:]]", "", MessyPostcode)
MessyPostcode <- toupper(MessyPostcode)
cln_str <- MessyPostcode
return(cln_str)
}
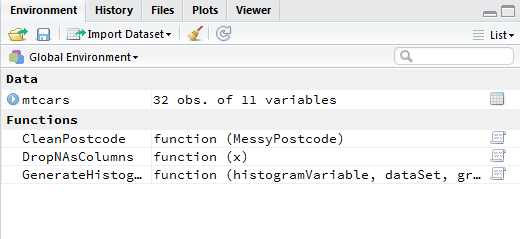
When I run the first file, the objects are available in the global environment:
There are some other function in the file but they are not relevant to the described problem.
Nevertheless the RStudio sees the object as not available in scope, as illustrated by the yellow triangle next to the code:
Question
Is there a way to make RStudio stop doing that. Maybe changing something to the source command? I tried local = TRUE and got the same thing. The code works with no problems, I just find it annoying.
The report was generated on the version 0.99.491 of RStudio.