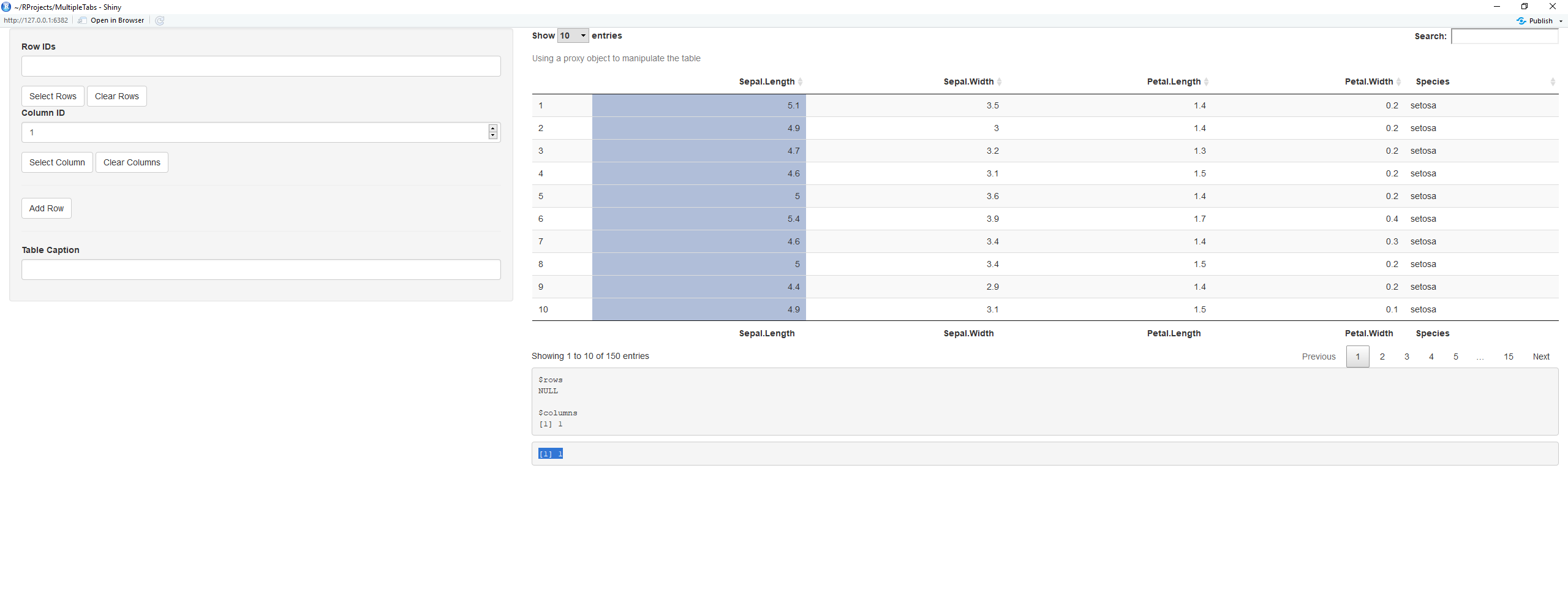
I have been working on trying to grasp how to make DT::dataTableProxy Using DT in Shiny work within my app, but I cannot get passed returning the selected columns.
Following the example here, I attempted to modify the code to print descriptive statistics (using pastecs). But the only thing that renders is the literal column selected.
#modified UI, adding another verbatimTextOutput
ui =
...
verbatimTextOutput('foo2')
server =
...
output$foo2= renderPrint({
x<-input$foo_columns_selected
stat.desc(x)
})
What I would like to do, is obtain the values of the selected column, and run the function
stat.desc. In the above example, it would be ran on column 2 (Sepal.Width), and render the descriptive statistics. Like this:
Going forward, I would like to have stat.desc performed on a rendered DataTable from multiple selectInputs. But..one step at a time. I want to grasp first how to get the actual values to perform functions on.
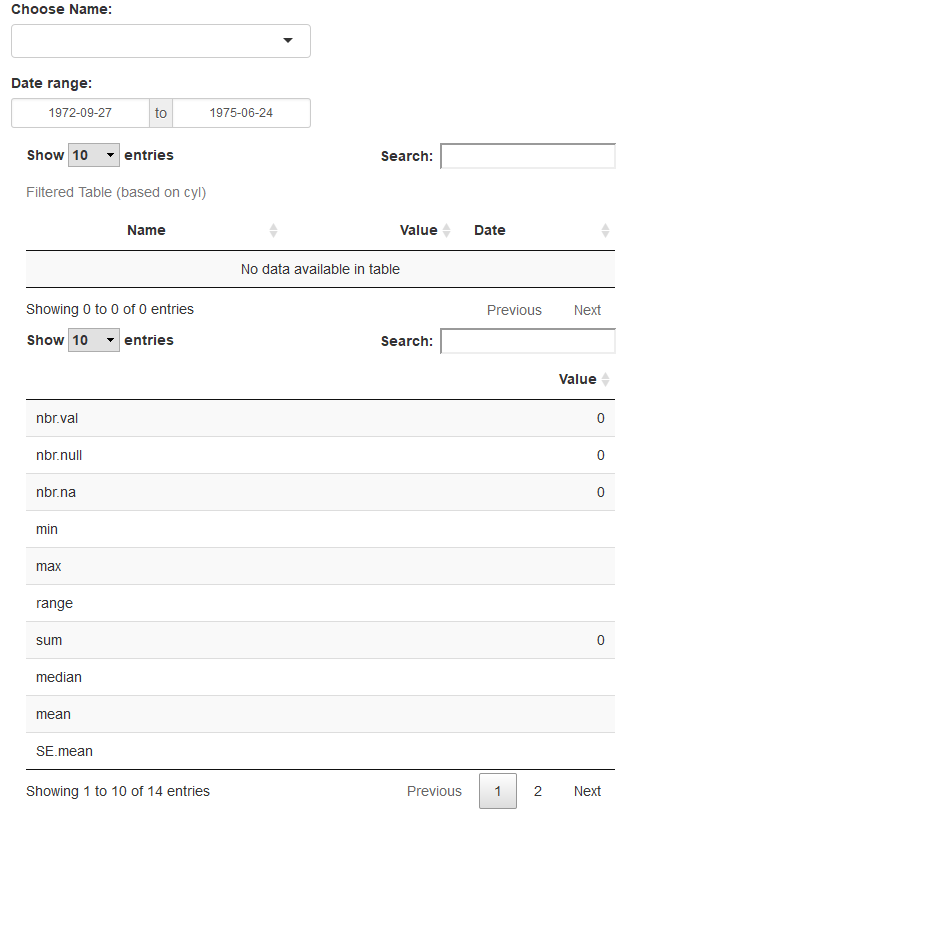
Update!
So, I think i figured out how to successfully accomplish what I needed. Would love if a fellow user could validate that it is working properly:
Updated script:
require(DT)
library(dplyr)
library(tibble)
ui<-
fluidPage(
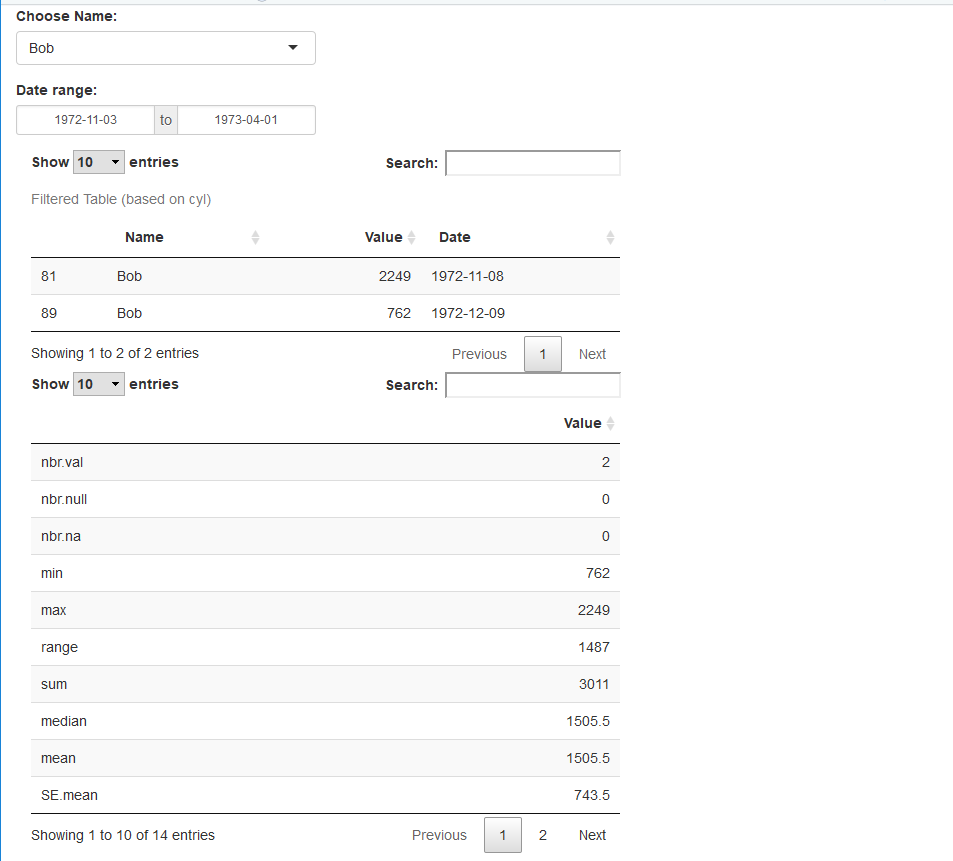
selectInput('obj','Choose Name:', choices = c('',my.data$Name), selectize = TRUE),
dateRangeInput('daterange',"Date range:",
start= min(my.data$Date),
end = max(my.data$Date)),
mainPanel(
dataTableOutput('filteredTable'),
dataTableOutput('filteredTable2'),
tableOutput('table')
)
)
server<-function(input,output, session){
filteredTable_data <- reactive({
my.data %>% rownames_to_column() %>% ##dplyr's awkward way to preserve rownames
filter(., Name == input$obj) %>%
filter(., between(Date ,input$daterange[1], input$daterange[2])) %>%
column_to_rownames()
})
##explicit assignment to output ID
DT::dataTableOutput("filteredTable")
output$filteredTable <- DT::renderDataTable({
datatable(
filteredTable_data(),
selection = list(mode = "multiple"),
caption = "Filtered Table (based on cyl)"
)
})
filteredTable_selected <- reactive({
ids <- input$filteredTable_rows_all
filteredTable_data()[sort(ids),] ##sort index to ensure orig df sorting
})
##anonymous
output$filteredTable2<-DT::renderDataTable({
x<-filteredTable_selected() %>% select(starts_with("Value"))
x<-as.data.frame(stat.desc(x))
datatable(
x)
})
}
shinyApp(ui, server)
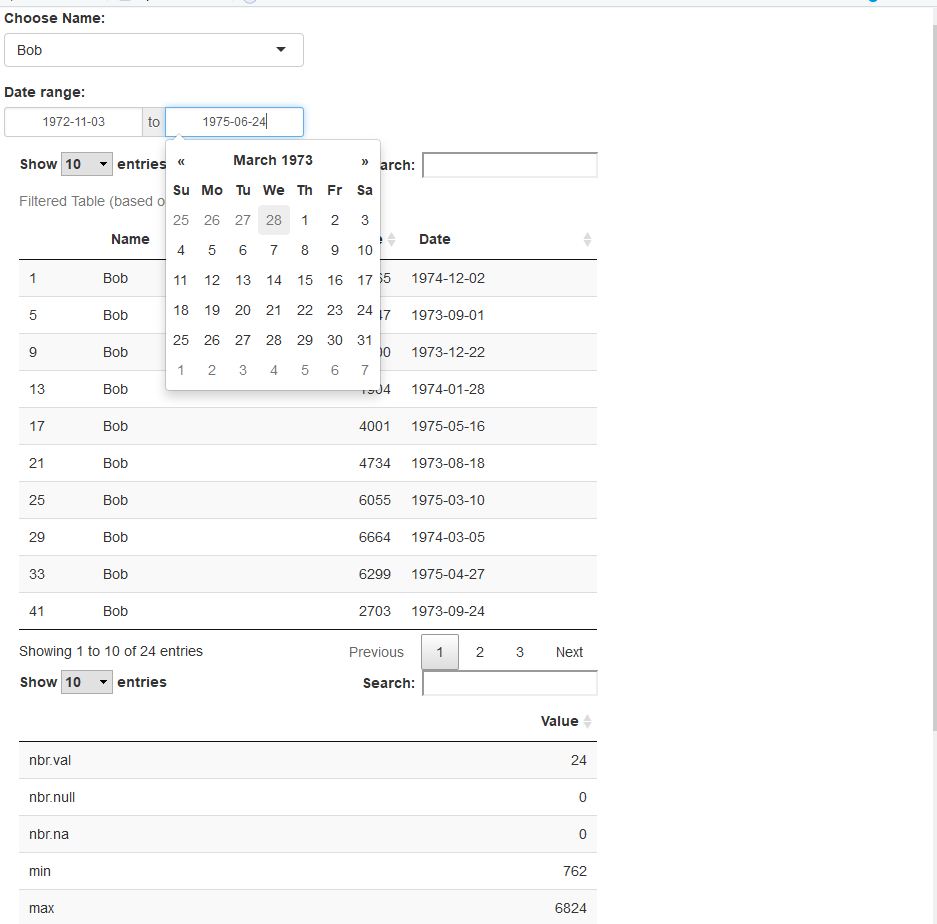
Result:
The following answers helped immensely: Reading objects from shiny output object not allowed? & How do I get the data from the selected rows of a filtered datatable (DT)?