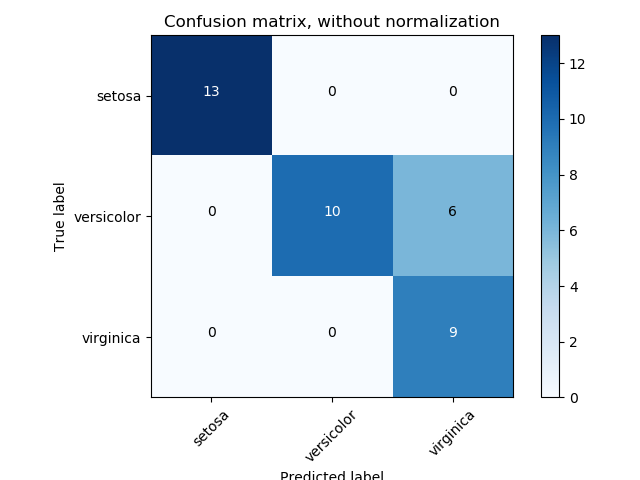
I have a set of data with known labels. I want to try clustering and see if I can get the same clusters given by known labels. To measure the accuracy, I need to get something like a confusion matrix.
I know I can get a confusion matrix easily for a test set of a classification problem. I already tried that like this.
However, it can't be used for clustering as it expected both columns and rows to have the same set of labels, which makes sense for a classification problem. But for a clustering problem what I expect is something like this.
Rows - Actual labels
Columns - New cluster names (i.e. cluster-1, cluster-2 etc.)
Is there a way to do this?
Edit: Here are more details.
In sklearn.metrics.confusion_matrix, it expects y_test and y_pred to have the same values, and labels to be the labels of those values.
That's why it gives a matrix which has the same labels for both rows and columns like this.
But in my case (KMeans Clustering), the real values are Strings and estimated values are numbers (i.e. cluster number)
Therefore, if I call confusion_matrix(y_true, y_pred) it gives below error.
ValueError: Mix of label input types (string and number)
This is the real problem. For a classification problem, this makes sense. But for a clustering problem, this restriction shouldn't be there, because real label names and new cluster names don't need to be the same.
With this, I understand I'm trying to use a tool, which is supposed to be used for classification problems, for a clustering problem. So, my question is, is there a way I can get such a matrix for may clustered data.
Hope the question is now clearer. Please let me know if it isn't.