I can create a tree with Rpart using the Kyphosis data set which is part of base R:
fit <- rpart(Kyphosis ~ Age + Number + Start,
method="class", data=kyphosis)
printcp(fit)
plot(fit, uniform=TRUE,main="Classification Tree for Kyphosis")
text(fit, use.n=TRUE, all=TRUE, cex=.8)
This is what the tree looks like:
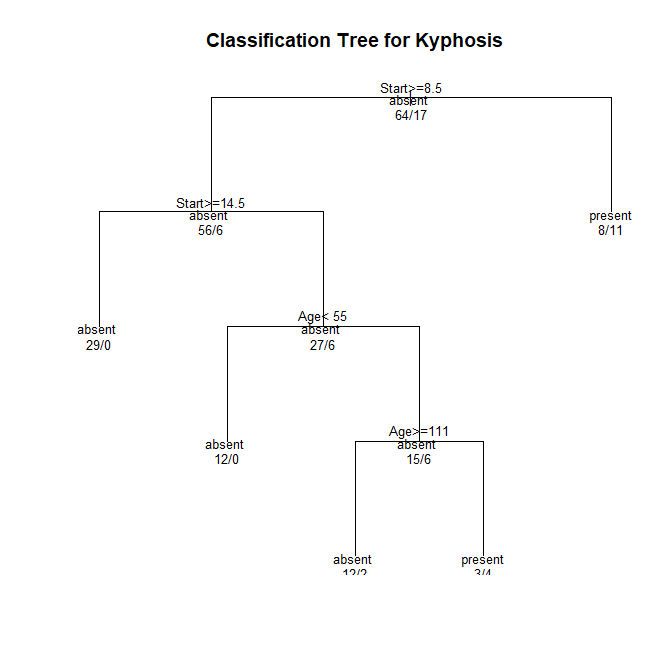
Now to visualize the tree better I want to make use of a sankey diagram using plotly. To create a sankey diagram in plotly one has to do the following:
library(plotly)
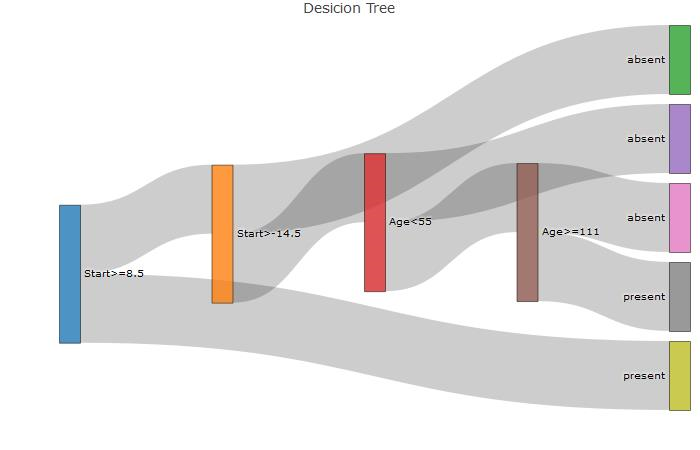
nodes=c("Start>=8.5","Start>-14.5","absent",
"Age<55","absent","Age>=111","absent","present","present")
p <- plot_ly(
type = "sankey",
orientation = "h",
node = list(
label = nodes,
pad = 10,
thickness = 20,
line = list(
color = "black",
width = 0.5
)
),
link = list(
source = c(0,1,1,3,3,5,5,0),
target = c(1,2,3,4,5,6,7,8),
value = c(1,1,1,1,1,1,1,1)
)
) %>%
layout(
title = "Desicion Tree",
font = list(
size = 10
)
)
p
This creates a sankey diagram corresponding to the tree(hard coded). The three necessary vectors needed are 'source','target','value' and looks as follows:
Hard coded sankey diagram:
My problem is using the rpart object 'fit' I can't seem to easily obtain a vector to produce the required 'source','target' and 'value' vectors for plotly.
fit$frame and fit$splits contains some of the information but it's difficult to aggregate them or use together. Using the print function on the fit object produces the needed information but I don't want to do text editing to obtain it.
print(fit)
Output:
1) root 81 17 absent (0.79012346 0.20987654)
2) Start>=8.5 62 6 absent (0.90322581 0.09677419)
4) Start>=14.5 29 0 absent (1.00000000 0.00000000) *
5) Start< 14.5 33 6 absent (0.81818182 0.18181818)
10) Age< 55 12 0 absent (1.00000000 0.00000000) *
11) Age>=55 21 6 absent (0.71428571 0.28571429)
22) Age>=111 14 2 absent (0.85714286 0.14285714) *
23) Age< 111 7 3 present (0.42857143 0.57142857) *
3) Start< 8.5 19 8 present (0.42105263 0.57894737) *
So is there an easy way to use a rpart object to obtain those 3 vectors for plotly to produce a sankey diagram? This plot will be used in a web app so plotly must be used since we already have javascript that correponds to it and it must easily be reusable to be applied to various datasets.

fitobject? – Sayette