I'm trying to calculate the weighted topological overlap for an adjacency matrix but I cannot figure out how to do it correctly using numpy. The R function that does the correct implementation is from WGCNA (https://www.rdocumentation.org/packages/WGCNA/versions/1.67/topics/TOMsimilarity). The formula for computing this (I THINK) is detailed in equation 4 which I believe is correctly reproduced below.
Does anyone know how to implement this correctly so it reflects the WGCNA version?
Yes, I know about rpy2 but I'm trying to go lightweight on this if possible.
For starters, my diagonal is not 1 and the values have no consistent error from the original (e.g. not all off by x).
When I computed this in R, I used the following:
> library(WGCNA, quiet=TRUE)
> df_adj = read.csv("https://pastebin.com/raw/sbAZQsE6", row.names=1, header=TRUE, check.names=FALSE, sep="\t")
> df_tom = TOMsimilarity(as.matrix(df_adj), TOMType="unsigned", TOMDenom="min")
# ..connectivity..
# ..matrix multiplication (system BLAS)..
# ..normalization..
# ..done.
# I've uploaded it to this url: https://pastebin.com/raw/HT2gBaZC
I'm not sure where my code is incorrect. The source code for the R version is here but it's using C backend scripts? which is very difficult for me interpret.
Here is my implementation in Python :
import pandas as pd
import numpy as np
from sklearn.datasets import load_iris
def get_iris_data():
iris = load_iris()
# Iris dataset
X = pd.DataFrame(iris.data,
index = [*map(lambda x:f"iris_{x}", range(150))],
columns = [*map(lambda x: x.split(" (cm)")[0].replace(" ","_"), iris.feature_names)])
y = pd.Series(iris.target,
index = X.index,
name = "Species")
return X, y
# Get data
X, y = get_iris_data()
# Create an adjacency network
# df_adj = np.abs(X.T.corr()) # I've uploaded this part to this url: https://pastebin.com/raw/sbAZQsE6
df_adj = pd.read_csv("https://pastebin.com/raw/sbAZQsE6", sep="\t", index_col=0)
A_adj = df_adj.values
# Correct TOM from WGCNA for the A_adj
# See above for code
# https://www.rdocumentation.org/packages/WGCNA/versions/1.67/topics/TOMsimilarity
df_tom__wgcna = pd.read_csv("https://pastebin.com/raw/HT2gBaZC", sep="\t", index_col=0)
# My attempt
A = A_adj.copy()
dimensions = A.shape
assert dimensions[0] == dimensions[1]
d = dimensions[0]
# np.fill_diagonal(A, 0)
# Equation (4) from http://dibernardo.tigem.it/files/papers/2008/zhangbin-statappsgeneticsmolbio.pdf
A_tom = np.zeros_like(A)
for i in range(d):
a_iu = A[i]
k_i = a_iu.sum()
for j in range(i+1, d):
a_ju = A[:,j]
k_j = a_ju.sum()
l_ij = np.dot(a_iu, a_ju)
a_ij = A[i,j]
numerator = l_ij + a_ij
denominator = min(k_i, k_j) + 1 - a_ij
w_ij = numerator/denominator
A_tom[i,j] = w_ij
A_tom = (A_tom + A_tom.T)
There is a package called GTOM (https://github.com/benmaier/gtom) but it is not for weighted adjacencies. The author of GTOM also took a look at this problem (which a much more sophisticated/efficient NumPy implementation but it's still not producing the expected results).
Does anyone know how to reproduce the WGCNA implementation?
EDIT: 2019.06.20
I've adapted some of the code from @scleronomic and @benmaier with credits in the doc string. The function is available in soothsayer from v2016.06 and on. Hopefully this will allow people to use topological overlap in Python easier instead of only being able to use R.
https://github.com/jolespin/soothsayer/blob/master/soothsayer/networks/networks.py
import numpy as np
import soothsayer as sy
df_adj = sy.io.read_dataframe("https://pastebin.com/raw/sbAZQsE6")
df_tom = sy.networks.topological_overlap_measure(df_adj)
df_tom__wgcna = sy.io.read_dataframe("https://pastebin.com/raw/HT2gBaZC")
np.allclose(df_tom, df_tom__wgcna)
# True

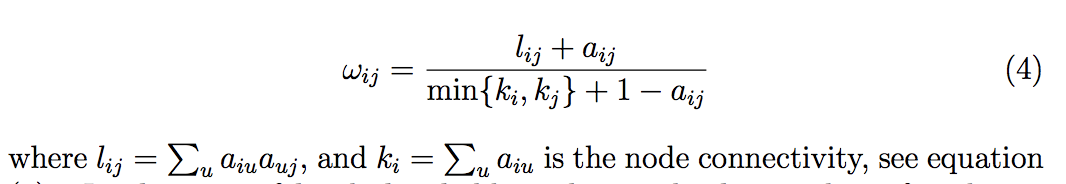

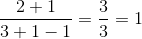
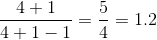
Lmatrix incorrectly ha ah man. – Apophyge