I would like to:
Reclassify the raster
rasinto nine classes usingreclassifyProvide a colorkey with values written beside each colour (see sample plot below). The colorkey should not be split, as in the sample plot, but rather shown as a single colour ramp with 9 colours.
Display the data using
levelplotfunction (this I can do)
Thanks for your help. AEZ
library(rasterVis)
Please use these colors:
col <- colorRampPalette(c("yellow4", "yellow", "orange", "red3", "darkred"))
levelplot(reclassras, col.regions = col)
Below is the dput of my raster layer:
ras=new("RasterLayer"
, file = new(".RasterFile"
, name = ""
, datanotation = "FLT4S"
, byteorder = "little"
, nodatavalue = -Inf
, NAchanged = FALSE
, nbands = 1L
, bandorder = "BIL"
, offset = 0L
, toptobottom = TRUE
, blockrows = 4L
, blockcols = 230L
, driver = ""
, open = FALSE
)
, data = new(".SingleLayerData"
, values = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, 0.693928846555399, 0.788672037295368, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, 0.63077312659247, 0.646355947246998, 0.877268680738184,
1.18198745233929, 1.17243141777504, 1.16780606010241, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 0.796574503892096, 0.619565545705973, 0.548990505935034,
0.595313378137405, 0.661379946396662, 0.780858688754648, 1.32843871685037,
1.3513059907622, 1.47504896477768, 1.33220740188043, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
1.02493198234592, 0.871180733341381, 0.588561527602423, 0.495423050558386,
0.398352388696117, 0.609004794796874, 0.757493518742196, 0.968962672868491,
1.3989459779932, 1.66210327179263, 1.4993019355018, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.605206815154586,
0.542575959157491, 0.509368162877491, 0.461856452801692, 0.543152629728101,
0.630122267439296, 0.70810504838076, 0.788478557501378, 0.918580431438071,
0.925176992886461, 0.712139151650403, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.523779588981426, 0.497759414675531,
0.539521493581999, 0.472379195067506, 0.503370029793095, 0.549265480778097,
0.570528210136693, 0.710933566259782, 0.726885712871992, 0.755087833088479,
0.692253167927366, 0.632483742020028, 0.572268262363865, 0.438810629423197,
0.495519373283201, 0.461307895792857, 0.413954466042644, 0.368257331706444,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.777296738735527,
0.750850589920605, 0.513284076851117, 0.491285172798063, 0.555785245510632,
0.611268469163931, 0.813189823687993, 0.756674620262126, 0.872121251992992,
0.613545590220432, 0.481851214001446, 0.496306521393404, 0.437621615262168,
0.339745859876615, 0.303638244461432, 0.311408628670555, 0.307152533981348,
0.31488062771456, 0.324014538449246, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, 0.682358704297568, 0.706551558881091, 0.502018311967365,
0.456684326929721, 0.473559918237202, 0.463389646751083, 0.441190726838794,
0.439217524749229, 0.584294316557062, 0.267633271553189, 0.256528025697032,
0.337787793432092, 0.330618969832041, 0.13803984358201, 0.157341965515659,
0.25899384267949, 0.217328457675264, 0.242480963818602, 0.272211051701126,
0.281644918669069, NA, NA, NA, NA, NA, NA, NA, NA, 0.414616765967516,
0.460428280883944, 0.439029889808256, 0.373948128082835, 0.352871572842356,
0.357951354005567, 0.328436868901166, 0.203201513096291, 0.141472917108049,
0.269096407445964, 0.15745212479733, 0.135179353417107, 0.219985246649717,
0.283005986377796, 0.209135929188723, 0.195612120899385, 0.163726230506747,
0.117276078938131, 0.169752099589576, 0.20586142557315, 0.244209631644345,
NA, NA, NA, NA, NA, 0.214238769725864, 0.208010897733397, 0.222208413054847,
0.212686463354896, 0.281732716326432, 0.300708262469288, 0.223204487650159,
0.200983714892431, 0.156651839437054, 0.224474592578023, 0.145079279318955,
0.137658404085203, 0.196204204670511, 0.157445860015357, 0.205907596393374,
0.250666664848486, 0.277186662044126, 0.265355210400489, 0.241808654124976,
0.199713317093066, 0.143711490248281, 0.0842822747178765, NA,
NA, NA, NA, 0.264092813395827, NA, 0.212441257972578, 0.189725290483582,
0.168955415365627, 0.158197543230828, 0.147705597579244, 0.157228804810736,
0.211249178305103, 0.222557582448186, 0.215124944446001, 0.210152099165574,
0.224764230646078, 0.192500267075076, 0.174923484036373, 0.121073670585186,
0.140937204004106, 0.213437474831745, 0.248947340425651, 0.265571783211699,
0.266099597629565, 0.254099129610638, 0.225777564711763, 0.178602219399,
NA, NA, NA, 0.245068225382459, 0.250977120426067, 0.255618295682899,
0.215763680642785, 0.192426673071868, 0.15695069948531, 0.113984884275593,
0.132801862169494, 0.166848777379759, 0.115940552527949, 0.157310632213977,
0.227022907248424, 0.239855727768449, 0.239340572413914, 0.231675634554819,
0.215388496476701, 0.185506053849589, 0.104971870404521, 0.112957653991221,
0.184468284077724, 0.222974948120878, 0.234747755547275, 0.240936436264378,
0.246124530363775, 0.242871371021741, NA, NA, NA, NA, 0.237419647069358,
0.238880206380025, 0.231523511204313, 0.179362897056799, 0.16381508377734,
0.133186183020653, 0.0908146529276369, 0.141301676274407, 0.17155039695241,
0.176667304881711, 0.192368230921037, 0.21608936753556, 0.226766198129308,
0.221609735358686, 0.196435659994756, 0.18030618027393, 0.178648959768383,
0.131173639965412, 0.0673928945097901, 0.144030029855376, 0.183644454323076,
0.164135425763568, 0.177071512975595, 0.218027665373501, 0.24020227724051,
NA, NA, NA, NA, 0.222719673381422, 0.203163737836875, 0.193893900175302,
0.173629261933378, 0.118612596801245, 0.120337304279883, 0.132270235949265,
0.145957944294342, 0.158947316774427, 0.171811369315789, 0.178481215022077,
0.182769116915529, 0.193576475278923, 0.187526186900056, 0.116642925509998,
0.0810130524927432, 0.155361056173399, 0.148275096869829, 0.110248050354185,
0.150980011296809, 0.149035884104822, 0.0943587027445095, NA,
NA, NA, NA, NA, NA, NA, NA, NA, 0.155642765186448, 0.165539811602326,
0.138858875725954, 0.115966143137369, 0.110762436834703, 0.116558297180592,
0.109461897495481, 0.137020886546595, 0.130569369589671, 0.106845763992692,
0.139732412594499, 0.167204357520135, 0.121743769239998, 0.0978888268526515,
0.161034204706105, 0.182178539628265, 0.179821789305561, 0.183479109899686,
0.160540641903203, 0.112272216175425, NA, NA, NA, NA, NA, NA,
NA, NA, NA, 0.163697841087448, 0.17328205868045, 0.160040552656876,
0.134655975429996, 0.12409849785916, 0.107880400167541, 0.0924636872864672,
0.103225290747982, 0.0767026155342135, 0.0661780888537736, 0.112966312114504,
0.16666404802081, 0.172067566098574, 0.172700784074864, 0.192001569629986,
0.206098509476636, 0.210359965210036, 0.209377663322926, 0.197655864616846,
0.180297988050893, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.181675191082483,
0.176935554759326, 0.16390655974363, 0.154194456563158, 0.142896144640483,
0.133243173249838, 0.114015642056713, 0.0928738807683648, 0.116223803463035,
0.146600649670514, 0.180171520048675, 0.197023191340036, 0.203375499409087,
NA, NA, NA, NA, 0.220951932247611, 0.216562385948588, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.176899006540279,
0.169734384672319, 0.164263225996517, 0.143295516102167, 0.13599420407605,
NA, 0.177143713627865, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 0.189498742688049,
0.185499135750397, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA)
, offset = 0
, gain = 1
, inmemory = TRUE
, fromdisk = FALSE
, isfactor = FALSE
, attributes = list()
, haveminmax = TRUE
, min = 0.0661780888537736
, max = 1.66210327179263
, band = 1L
, unit = ""
, names = "myfile"
)
, legend = new(".RasterLegend"
, type = character(0)
, values = logical(0)
, color = logical(0)
, names = logical(0)
, colortable = logical(0)
)
, title = character(0)
, extent = new("Extent"
, xmin = 73.39696
, xmax = 96.59696
, ymin = 33.98298
, ymax = 49.18298
)
, rotated = FALSE
, rotation = new(".Rotation"
, geotrans = numeric(0)
, transfun = function ()
NULL
)
, ncols = 29L
, nrows = 19L
, crs = new("CRS"
, projargs = "+proj=longlat +a=6378140 +b=6356755.288157528 +no_defs"
)
, history = list()
, z = list()
)

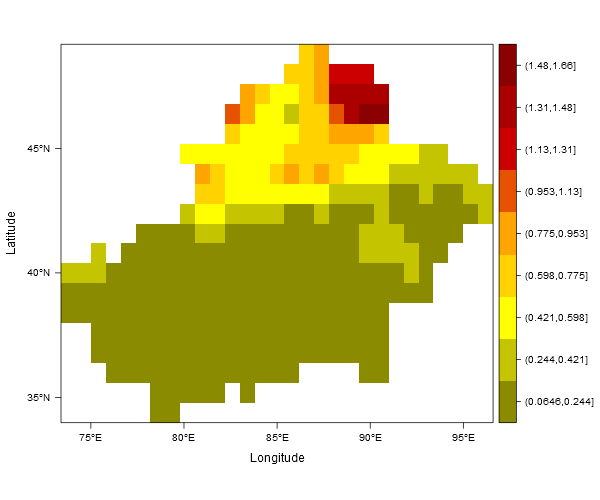
dputdoesn't work so well for rasters (you should always check that the code you provide runs in a clean R session - it doesn't in this case). You could replace your whole example raster withras <- raster(matrix(runif(100), nrow=10)). Also, I don't understand your second point (colorkey should be "joined"). – Furness